CYCLIN-DEPENDENT KINASE G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis
- PMID: 23404887
- PMCID: PMC3608783
- DOI: 10.1105/tpc.112.107896
CYCLIN-DEPENDENT KINASE G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis
Abstract
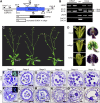
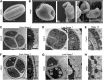
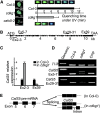
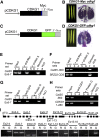
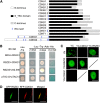
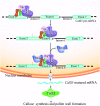
Arabidopsis thaliana CYCLIN-DEPEDENT KINASE G1 (CDKG1) belongs to the family of cyclin-dependent protein kinases that were originally characterized as cell cycle regulators in eukaryotes. Here, we report that CDKG1 regulates pre-mRNA splicing of CALLOSE SYNTHASE5 (CalS5) and, therefore, pollen wall formation. The knockout mutant cdkg1 exhibits reduced male fertility with impaired callose synthesis and abnormal pollen wall formation. The sixth intron in CalS5 pre-mRNA, a rare type of intron with a GC 5' splice site, is abnormally spliced in cdkg1. RNA immunoprecipitation analysis suggests that CDKG1 is associated with this intron. CDKG1 contains N-terminal Ser/Arg (RS) motifs and interacts with splicing factor Arginine/Serine-Rich Zinc Knuckle-Containing Protein33 (RSZ33) through its RS region to regulate proper splicing. CDKG1 and RS-containing Zinc Finger Protein22 (SRZ22), a splicing factor interacting with RSZ33 and U1 small nuclear ribonucleoprotein particle (snRNP) component U1-70k, colocalize in nuclear speckles and reside in the same complex. We propose that CDKG1 is recruited to U1 snRNP through RSZ33 to facilitate the splicing of the sixth intron of CalS5.
Figures

Similar articles
-
AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis.Plant Physiol. 2013 Jun;162(2):720-31. doi: 10.1104/pp.113.214940. Epub 2013 Apr 11. Plant Physiol. 2013. PMID: 23580594 Free PMC article.
-
The cyclin-dependent kinase G group defines a thermo-sensitive alternative splicing circuit modulating the expression of Arabidopsis ATU2AF65A.Plant J. 2018 Jun;94(6):1010-1022. doi: 10.1111/tpj.13914. Epub 2018 May 10. Plant J. 2018. PMID: 29602264 Free PMC article.
-
Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleoprotein-specific proteins suggest their involvement in pre-mRNA Splicing.J Biol Chem. 2004 Aug 6;279(32):33890-8. doi: 10.1074/jbc.M400270200. Epub 2004 May 27. J Biol Chem. 2004. PMID: 15166240
-
Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge.Trends Biochem Sci. 1997 Apr;22(4):132-7. doi: 10.1016/s0968-0004(97)01018-9. Trends Biochem Sci. 1997. PMID: 9149533 Review.
-
How Are Short Exons Flanked by Long Introns Defined and Committed to Splicing?Trends Genet. 2016 Oct;32(10):596-606. doi: 10.1016/j.tig.2016.07.003. Epub 2016 Aug 6. Trends Genet. 2016. PMID: 27507607 Review.
Cited by
-
Overexpression of AtTTP affects ARF17 expression and leads to male sterility in Arabidopsis.PLoS One. 2015 Mar 30;10(3):e0117317. doi: 10.1371/journal.pone.0117317. eCollection 2015. PLoS One. 2015. PMID: 25822980 Free PMC article.
-
Microsporocytic ARF17 misexpression leads to an excess callose deposition and male sterility in Arabidopsis.Plant Mol Biol. 2025 Jan 17;115(1):18. doi: 10.1007/s11103-024-01549-3. Plant Mol Biol. 2025. PMID: 39821735
-
The Arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis.EMBO J. 2020 Feb 3;39(3):e101625. doi: 10.15252/embj.2019101625. Epub 2019 Sep 26. EMBO J. 2020. PMID: 31556459 Free PMC article.
-
Molecular Targets and Biological Functions of cAMP Signaling in Arabidopsis.Biomolecules. 2021 May 3;11(5):688. doi: 10.3390/biom11050688. Biomolecules. 2021. PMID: 34063698 Free PMC article.
-
Callose biosynthesis in Arabidopsis with a focus on pathogen response: what we have learned within the last decade.Ann Bot. 2014 Oct;114(6):1349-58. doi: 10.1093/aob/mcu120. Epub 2014 Jul 1. Ann Bot. 2014. PMID: 24984713 Free PMC article. Review.
References
-
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 - PubMed
-
- Ariizumi T., Hatakeyama K., Hinata K., Inatsugi R., Nishida I., Sato S., Kato T., Tabata S., Toriyama K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39: 170–181 - PubMed
-
- Ariizumi T., Toriyama K. (2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62: 437–460 - PubMed
-
- Bentley D. (1999). Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11: 347–351 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

