Generation and analysis of a complete mutant set for the Arabidopsis FT/TFL1 family shows specific effects on thermo-sensitive flowering regulation
- PMID: 23404901
- PMCID: PMC3617836
- DOI: 10.1093/jxb/ert036
Generation and analysis of a complete mutant set for the Arabidopsis FT/TFL1 family shows specific effects on thermo-sensitive flowering regulation
Abstract
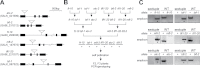
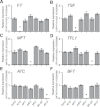
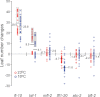
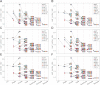
The FLOWERING LOCUS T (FT)/TERMINAL FLOWER 1 (TFL1) family proteins play an important role in the regulation of flowering time. In the Arabidopsis thaliana genome, there are six genes in the FT/TFL1 family. To determine how these FT/TFL1 family genes contribute to the regulation of flowering time, this study generated a comprehensive set of mutants (sixty-three multiple mutants in all combinations) of the FT/TFL1 family genes and analysed their flowering times at 23 and 16°C under long-day conditions. The analysis confirmed that FT and TFL1 are major determinants of flowering time under long-day conditions. At 23 °C, ft-10 tsf-1 mft-2 showed the latest flowering, whereas tfl1-20 atc-2 bft-2 showed the earliest flowering. Flowering occurred in the sextuple mutants. Introduction of tsf-1 led to reduced sensitivity to ambient temperature change. Introduction of tfl1-20 caused a stronger effect in accelerating flowering time at 16 °C than at 23 °C. Overexpression of miR156 did not block flowering of sextuple mutants, suggesting that there is a pathway to induce flowering independent of the FT/TFL1 pathway and miR156 pathway. This study proposes that this mutant population will be useful in further investigation of the functions of the FT/TFL1 family genes in plant development.
Figures


References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 - PubMed
-
- Blazquez MA, Ahn JH, Weigel D. 2003. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nature Genetics 33, 168–171 - PubMed
-
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. 1997. Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

