Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer
- PMID: 23405235
- PMCID: PMC3566108
- DOI: 10.1371/journal.pone.0055910
Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer
Abstract
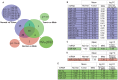
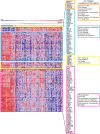
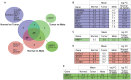
Triple negative breast cancer (TNBC) is a heterogeneous disease at the molecular, pathologic and clinical levels. To stratify TNBCs, we determined microRNA (miRNA) expression profiles, as well as expression profiles of a cancer-focused mRNA panel, in tumor, adjacent non-tumor (normal) and lymph node metastatic lesion (mets) tissues, from 173 women with TNBCs; we linked specific miRNA signatures to patient survival and used miRNA/mRNA anti-correlations to identify clinically and genetically different TNBC subclasses. We also assessed miRNA signatures as potential regulators of TNBC subclass-specific gene expression networks defined by expression of canonical signal pathways.Tissue specific miRNAs and mRNAs were identified for normal vs tumor vs mets comparisons. miRNA signatures correlated with prognosis were identified and predicted anti-correlated targets within the mRNA profile were defined. Two miRNA signatures (miR-16, 155, 125b, 374a and miR-16, 125b, 374a, 374b, 421, 655, 497) predictive of overall survival (P = 0.05) and distant-disease free survival (P = 0.009), respectively, were identified for patients 50 yrs of age or younger. By multivariate analysis the risk signatures were independent predictors for overall survival and distant-disease free survival. mRNA expression profiling, using the cancer-focused mRNA panel, resulted in clustering of TNBCs into 4 molecular subclasses with different expression signatures anti-correlated with the prognostic miRNAs. Our findings suggest that miRNAs play a key role in triple negative breast cancer through their ability to regulate fundamental pathways such as: cellular growth and proliferation, cellular movement and migration, Extra Cellular Matrix degradation. The results define miRNA expression signatures that characterize and contribute to the phenotypic diversity of TNBC and its metastasis.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

