Helper T-cell identity and evolution of differential transcriptomes and epigenomes
- PMID: 23405893
- PMCID: PMC3577092
- DOI: 10.1111/imr.12037
Helper T-cell identity and evolution of differential transcriptomes and epigenomes
Abstract
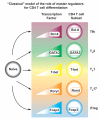
CD4(+) T cells are critical for the elimination of an immense array of microbial pathogens. Among the ways they accomplish this task is to generate progeny with specialized, characteristic patterns of gene expression. From this perspective, helper cells can be viewed as pluripotent precursors that adopt distinct cell fates. Although there are aspects of helper cell differentiation that can be modeled as a classic cell fate commitment, CD4(+) T cells also maintain considerable flexibility in their transcriptional program. This makes sense in terms of host defense, but raises the question of how these remarkable cells balance both these requirements, a high degree of specific gene expression and the capacity for plasticity. In this review, we discuss recent advances in our understanding of CD4(+) T-cell specification, focusing on how genomic perspectives have influenced our views of these processes. The relative contributions of sensors of the cytokine milieu, especially the signal transducer and activator of transcription family transcription factors, 'master regulators', and other transcription factors are considered as they relate to the helper cell transcriptome and epigenome.
Published 2013. This article is a U.S. Government work and is in the public domain in the USA.
Figures


References
-
- Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33:12–24. - PubMed
-
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. - PubMed
-
- Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

