Alzheimer's Aβ42 and Aβ40 peptides form interlaced amyloid fibrils
- PMID: 23406382
- PMCID: PMC3716832
- DOI: 10.1111/jnc.12202
Alzheimer's Aβ42 and Aβ40 peptides form interlaced amyloid fibrils
Abstract
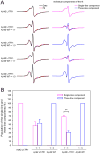
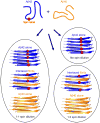
Deposition of amyloid β (Aβ) in the brain is a pathological hallmark of Alzheimer's disease. There are two major isoforms of Aβ: the 42-residue Aβ42 and the 40-residue Aβ40. The only difference between Aβ42 and Aβ40 is that Aβ42 has two extra residues at the C-terminus. The amyloid plaques in Alzheimer's brains consist of mostly Aβ42 and some plaques contain only Aβ42, even though Aβ40 concentration is several-fold more than Aβ42. Using electron paramagnetic resonance, we studied the formation of amyloid fibrils using a mixture of Aβ42 and Aβ40 in vitro. We show that Aβ42 and Aβ40 form mixed fibrils in an interlaced manner, although Aβ40 is not as efficient as Aβ42 in terms of being incorporated into Aβ42 fibrils. Our results suggest that both Aβ42 and Aβ40 would be present in amyloid plaques if in vivo aggregation of Aβ were similar to the in vitro process. Therefore, there must be some mechanisms that lead to the preferential deposition of Aβ42 at the extracellular space. Identifying such mechanisms may open new avenues for therapeutic interventions to treat Alzheimer's disease.
Keywords: electron paramagnetic resonance; protein aggre-gation; senile plaques; spin labeling.
© 2013 International Society for Neurochemistry.
Figures


References
-
- Agopian A, Guo Z. Structural origin of polymorphism for Alzheimer's amyloid-β fibrils. Biochem J. 2012;447:43–50. - PubMed
-
- Baker RT, Catanzariti AM, Karunasekara Y, Soboleva TA, Sharwood R, Whitney S, Board PG. Using deubiquitylating enzymes as research tools. Methods Enzymol. 2005;398:540–554. - PubMed
-
- Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. A new structural model of Aβ40 fibrils. J Am Chem Soc. 2011;133:16013–16022. - PubMed
-
- Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J Magn Reson, Ser A. 1996;120:155–189.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

