Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse
- PMID: 23406902
- PMCID: PMC4074262
- DOI: 10.1242/dev.084665
Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse
Abstract
The myogenic regulatory factor Myod and insulin-like growth factor 2 (Igf2) have been shown to interact in vitro during myogenic differentiation. In order to understand how they interact in vivo, we produced double-mutant mice lacking both the Myod and Igf2 genes. Surprisingly, these mice display neonatal lethality due to severe diaphragm atrophy. Alteration of diaphragm muscle development occurs as early as 15.5 days post-coitum in the double-mutant embryos and leads to a defect in the terminal differentiation of muscle progenitor cells. A negative-feedback loop was detected between Myod and Igf2 in embryonic muscles. Igf2 belongs to the imprinted H19-Igf2 locus. Molecular analyses show binding of Myod on a mesodermal enhancer (CS9) of the H19 gene. Chromatin conformation capture experiments reveal direct interaction of CS9 with the H19 promoter, leading to increased H19 expression in the presence of Myod. In turn, the non-coding H19 RNA represses Igf2 expression in trans. In addition, Igf2 also negatively regulates Myod expression, possibly by reducing the expression of the Srf transcription factor, a known Myod activator. In conclusion, Igf2 and Myod are tightly co-regulated in skeletal muscles and act in parallel pathways in the diaphragm, where they affect the progression of myogenic differentiation. Igf2 is therefore an essential player in the formation of a functional diaphragm in the absence of Myod.
Figures
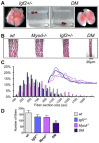

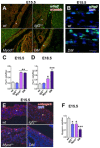
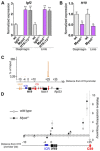

References
-
- Braem C., Recolin B., Rancourt R. C., Angiolini C., Barthès P., Branchu P., Court F., Cathala G., Ferguson-Smith A. C., Forné T. (2008). Genomic matrix attachment region and chromosome conformation capture quantitative real time PCR assays identify novel putative regulatory elements at the imprinted Dlk1/Gtl2 locus. J. Biol. Chem. 283, 18612–18620 - PubMed
-
- Bröhl D., Vasyutina E., Czajkowski M. T., Griger J., Rassek C., Rahn H. P., Purfürst B., Wende H., Birchmeier C. (2012). Colonization of the satellite cell niche by skeletal muscle progenitor cells depends on Notch signals. Dev. Cell 23, 469–481 - PubMed
-
- Buckingham M. (2007). Skeletal muscle progenitor cells and the role of Pax genes. C. R. Biol. 330, 530–533 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

