Remodeling of three-dimensional organization of the nucleus during terminal keratinocyte differentiation in the epidermis
- PMID: 23407401
- PMCID: PMC4135477
- DOI: 10.1038/jid.2013.66
Remodeling of three-dimensional organization of the nucleus during terminal keratinocyte differentiation in the epidermis
Abstract
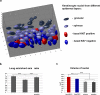
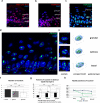
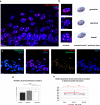
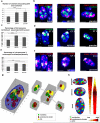
The nucleus of epidermal keratinocytes (KCs) is a complex and highly compartmentalized organelle, whose structure is markedly changed during terminal differentiation and transition of the genome from a transcriptionally active state seen in the basal and spinous epidermal cells to a fully inactive state in the keratinized cells of the cornified layer. Here, using multicolor confocal microscopy, followed by computational image analysis and mathematical modeling, we demonstrate that in normal mouse footpad epidermis, transition of KCs from basal epidermal layer to the granular layer is accompanied by marked differences in nuclear architecture and microenvironment including the following: (i) decrease in the nuclear volume; (ii) decrease in expression of the markers of transcriptionally active chromatin; (iii) internalization and decrease in the number of nucleoli; (iv) increase in the number of pericentromeric heterochromatic clusters; and (v) increase in the frequency of associations between the pericentromeric clusters, chromosomal territory 3, and nucleoli. These data suggest a role for nucleoli and pericentromeric heterochromatin clusters as organizers of nuclear microenvironment required for proper execution of gene expression programs in differentiating KCs, and provide important background information for further analyses of alterations in the topological genome organization seen in pathological skin conditions, including disorders of epidermal differentiation and epidermal tumors.
Figures
Comment in
-
Nuclear topology, epigenetics, and keratinocyte differentiation.J Invest Dermatol. 2013 Sep;133(9):2130-3. doi: 10.1038/jid.2013.261. J Invest Dermatol. 2013. PMID: 23949766
References
-
- Alcobia I, Dilao R, Parreira L. Spatial associations of centromeres in the nuclei of hematopoietic cells: evidence for cell-type-specific organizational patterns. Blood. 2000;95:1608–1615. - PubMed
-
- Aleixandre C, Miller DA, Mitchell AR, Warburton DA, Gersen SL, Disteche C, et al. p82H identifies sequences at every human centromere. Hum Genet. 1987;77:46–50. - PubMed
-
- Bazzi H, Fantauzzo KA, Richardson GD, Jahoda CA, Christiano AM. Transcriptional profiling of developing mouse epidermis reveals novel patterns of coordinated gene expression. Dev Dyn. 2007;236:961–970. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

