The affinity purification and characterization of ATP synthase complexes from mitochondria
- PMID: 23407638
- PMCID: PMC3603449
- DOI: 10.1098/rsob.120160
The affinity purification and characterization of ATP synthase complexes from mitochondria
Abstract
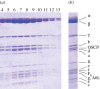
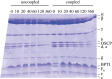
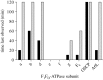
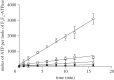
The mitochondrial F₁-ATPase inhibitor protein, IF₁, inhibits the hydrolytic, but not the synthetic activity of the F-ATP synthase, and requires the hydrolysis of ATP to form the inhibited complex. In this complex, the α-helical inhibitory region of the bound IF₁ occupies a deep cleft in one of the three catalytic interfaces of the enzyme. Its N-terminal region penetrates into the central aqueous cavity of the enzyme and interacts with the γ-subunit in the enzyme's rotor. The intricacy of forming this complex and the binding mode of the inhibitor endow IF₁ with high specificity. This property has been exploited in the development of a highly selective affinity procedure for purifying the intact F-ATP synthase complex from mitochondria in a single chromatographic step by using inhibitor proteins with a C-terminal affinity tag. The inhibited complex was recovered with residues 1-60 of bovine IF₁ with a C-terminal green fluorescent protein followed by a His-tag, and the active enzyme with the same inhibitor with a C-terminal glutathione-S-transferase domain. The wide applicability of the procedure has been demonstrated by purifying the enzyme complex from bovine, ovine, porcine and yeast mitochondria. The subunit compositions of these complexes have been characterized. The catalytic properties of the bovine enzyme have been studied in detail. Its hydrolytic activity is sensitive to inhibition by oligomycin, and the enzyme is capable of synthesizing ATP in vesicles in which the proton-motive force is generated from light by bacteriorhodopsin. The coupled enzyme has been compared by limited trypsinolysis with uncoupled enzyme prepared by affinity chromatography. In the uncoupled enzyme, subunits of the enzyme's stator are degraded more rapidly than in the coupled enzyme, indicating that uncoupling involves significant structural changes in the stator region.
Figures


References
-
- Walker JE. 1998. ATP synthesis by rotary catalysis. Angew. Chem. Int. Ed. 37, 5000–5011 10.1002/(SICI)1521-3773(19980918)37:17<2308::AID-ANIE2308>3.0.CO;2-W (doi:10.1002/(SICI)1521-3773(19980918)37:17<2308::AID-ANIE2308>3.0.CO;2-W) - DOI - PubMed
-
- Walker JE. 2012. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 41, 1–16 10.1042/BST20110773 (doi:10.1042/BST20110773) - DOI - PubMed
-
- Abrahams JP, Leslie AGW, Lutter R, Walker JE. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 10.1038/370621a0 (doi:10.1038/370621a0) - DOI - PubMed
-
- Bowler MW, Montgomery MG, Leslie AGW, Walker JE. 2006. How azide inhibits ATP hydrolysis by the F-ATPases. Proc. Natl Acad. Sci. USA 103, 8646–8649 10.1073/pnas.0602915103 (doi:10.1073/pnas.0602915103) - DOI - PMC - PubMed
-
- Bowler MW, Montgomery MG, Leslie AGW, Walker JE. 2007. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J. Biol. Chem. 282, 14 238–14 242 10.1074/jbc.M700203200 (doi:10.1074/jbc.M700203200) - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
