A family of diatom-like silicon transporters in the siliceous loricate choanoflagellates
- PMID: 23407828
- PMCID: PMC3574361
- DOI: 10.1098/rspb.2012.2543
A family of diatom-like silicon transporters in the siliceous loricate choanoflagellates
Abstract
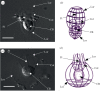
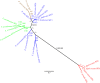
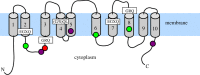
Biosilicification is widespread across the eukaryotes and requires concentration of silicon in intracellular vesicles. Knowledge of the molecular mechanisms underlying this process remains limited, with unrelated silicon-transporting proteins found in the eukaryotic clades previously studied. Here, we report the identification of silicon transporter (SIT)-type genes from the siliceous loricate choanoflagellates Stephanoeca diplocostata and Diaphanoeca grandis. Until now, the SIT gene family has been identified only in diatoms and other siliceous stramenopiles, which are distantly related to choanoflagellates among the eukaryotes. This is the first evidence of similarity between SITs from different eukaryotic supergroups. Phylogenetic analysis indicates that choanoflagellate and stramenopile SITs form distinct monophyletic groups. The absence of putative SIT genes in any other eukaryotic groups, including non-siliceous choanoflagellates, leads us to propose that SIT genes underwent a lateral gene transfer event between stramenopiles and loricate choanoflagellates. We suggest that the incorporation of a foreign SIT gene into the stramenopile or choanoflagellate genome resulted in a major metabolic change: the acquisition of biomineralized silica structures. This hypothesis implies that biosilicification has evolved multiple times independently in the eukaryotes, and paves the way for a better understanding of the biochemical basis of silicon transport through identification of conserved sequence motifs.
Figures

References
-
- Simkiss K, Wilbur KM. 1989. Biomineralization: cell biology and mineral deposition. San Diego, CA: Academic Press Inc
-
- Gröger C, Lutz K, Brunner E. 2008. Biomolecular self-assembly and its relevance in silica biomineralization. Cell Biochem. Biophys. 50, 23–3910.1007/s12013-007-9003-2 (doi:10.1007/s12013-007-9003-2) - DOI - DOI - PubMed
-
- Maldonado M, Carmona MC, Cruzado A. 1999. Decline in Mesozoic reef-building in sponges explained by silicon limitation. Nature 401, 785–78810.1038/44560 (doi:10.1038/44560). - DOI - DOI
-
- Hildebrand M, Higgins DR, Busser K, Volcani BE. 1993. Silicon-responsive cDNA clones isolated from the marine diatom Cylindrotheca fusiformis. Gene 132, 213–21810.1016/0378-1119(93)90198-C (doi:10.1016/0378-1119(93)90198-C) - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
