Non-catalytic participation of the Pin1 peptidyl-prolyl isomerase domain in target binding
- PMID: 23407864
- PMCID: PMC3571201
- DOI: 10.3389/fphys.2013.00018
Non-catalytic participation of the Pin1 peptidyl-prolyl isomerase domain in target binding
Abstract
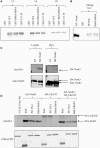
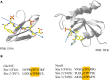
Pin1 is a phosphorylation-dependent peptidyl-prolyl isomerase (PPIase) that has the potential to add an additional level of regulation within protein kinase mediated signaling pathways. Furthermore, there is a mounting body of evidence implicating Pin1 in the emergence of pathological phenotypes in neurodegeneration and cancer through the isomerization of a wide variety of substrates at peptidyl-prolyl bonds where the residue preceding proline is a phosphorylated serine or threonine residue (i.e., pS/T-P motifs). A key step in this regulatory process is the interaction of Pin-1 with its substrates. This is a complex process since Pin1 is composed of two domains, the catalytic PPIase domain, and a type IV WW domain, both of which recognize pS/T-P motifs. The observation that the WW domain exhibits considerably higher binding affinity for pS/T-P motifs has led to predictions that the two domains may have distinct roles in mediating the actions of Pin1 on its substrates. To evaluate the participation of its individual domains in target binding, we performed GST pulldowns to monitor interactions between various forms of Pin1 and mitotic phospho-proteins that revealed two classes of Pin-1 interacting proteins, differing in their requirement for residues within the PPIase domain. From these observations, we consider models for Pin1-substrate interactions and the potential functions of the different classes of Pin1 interacting proteins. We also compare sequences that are recognized by Pin1 within its individual interaction partners to investigate the underlying basis for its different types of interactions.
Keywords: Pin1; WW domain; peptidyl-prolyl isomerase; peptidyl-prolyl isomerization; phosphorylation-dependent interactions.
Figures


Similar articles
-
Gears-In-Motion: The Interplay of WW and PPIase Domains in Pin1.Front Oncol. 2018 Oct 25;8:469. doi: 10.3389/fonc.2018.00469. eCollection 2018. Front Oncol. 2018. PMID: 30460195 Free PMC article. Review.
-
A tethering mechanism underlies Pin1-catalyzed proline cis-trans isomerization at a noncanonical site.Proc Natl Acad Sci U S A. 2025 May 27;122(21):e2414606122. doi: 10.1073/pnas.2414606122. Epub 2025 May 19. Proc Natl Acad Sci U S A. 2025. PMID: 40388619
-
The Peptidyl-Prolyl cis-trans isomerase, Pin1, associates with Protein Kinase C θ via a critical Phospho-Thr-Pro motif in the V3 regulatory domain.Front Immunol. 2023 Mar 8;14:1126464. doi: 10.3389/fimmu.2023.1126464. eCollection 2023. Front Immunol. 2023. PMID: 36969236 Free PMC article.
-
Neighboring phosphoSer-Pro motifs in the undefined domain of IRAK1 impart bivalent advantage for Pin1 binding.FEBS J. 2016 Dec;283(24):4528-4548. doi: 10.1111/febs.13943. Epub 2016 Nov 20. FEBS J. 2016. PMID: 27790836 Free PMC article.
-
The Multiple Roles of Peptidyl Prolyl Isomerases in Brain Cancer.Biomolecules. 2018 Oct 11;8(4):112. doi: 10.3390/biom8040112. Biomolecules. 2018. PMID: 30314361 Free PMC article. Review.
Cited by
-
Dissecting the functional behavior of the differentially phosphorylated prolyl isomerase, Pin1.Protein Sci. 2024 Sep;33(9):e5138. doi: 10.1002/pro.5138. Protein Sci. 2024. PMID: 39150071 Free PMC article.
-
Gears-In-Motion: The Interplay of WW and PPIase Domains in Pin1.Front Oncol. 2018 Oct 25;8:469. doi: 10.3389/fonc.2018.00469. eCollection 2018. Front Oncol. 2018. PMID: 30460195 Free PMC article. Review.
-
Activity-dependent isomerization of Kv4.2 by Pin1 regulates cognitive flexibility.Nat Commun. 2020 Mar 26;11(1):1567. doi: 10.1038/s41467-020-15390-x. Nat Commun. 2020. PMID: 32218435 Free PMC article.
-
Regulation of eukaryotic protein kinases by Pin1, a peptidyl-prolyl isomerase.Adv Biol Regul. 2023 Jan;87:100938. doi: 10.1016/j.jbior.2022.100938. Epub 2022 Nov 30. Adv Biol Regul. 2023. PMID: 36496344 Free PMC article.
-
Activity and Affinity of Pin1 Variants.Molecules. 2019 Dec 20;25(1):36. doi: 10.3390/molecules25010036. Molecules. 2019. PMID: 31861908 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

