Replication protein a links cell cycle progression and the onset of neurogenesis in Drosophila optic lobe development
- PMID: 23407946
- PMCID: PMC6619198
- DOI: 10.1523/JNEUROSCI.3357-12.2013
Replication protein a links cell cycle progression and the onset of neurogenesis in Drosophila optic lobe development
Abstract
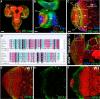
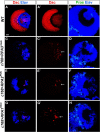
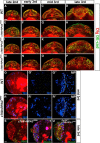
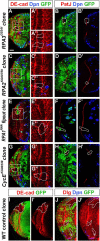
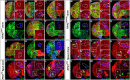
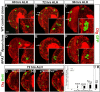
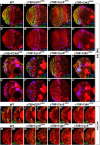
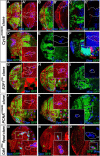
Stem cell self-renewal and differentiation must be carefully controlled during development and tissue homeostasis. In the Drosophila optic lobe, neuroepithelial cells first divide symmetrically to expand the stem cell population and then transform into asymmetrically dividing neuroblasts, which generate medulla neurons. The mechanisms underlying this cell fate transition are not well understood. Here, we show a crucial role of some cell cycle regulators in this transition. We find that loss of function in replication protein A (RPA), which consists of three highly conserved protein subunits and functions in DNA replication, leads to disintegration of the optic lobe neuroepithelium and premature differentiation of neuroepithelial cells into medulla neuroblasts. Clonal analyses of RPA loss-of-function alleles indicate that RPA is required to prevent neuroepithelial cells from differentiating into medulla neuroblasts. Inactivation of the core cell cycle regulators, including the G1/S regulators E2F1, Cyclin E, Cdk2, and PCNA, and the G2/M regulators Cyclin A, Cyclin B, and Cdk1, mimic RPA loss-of-function phenotypes, suggesting that cell cycle progression is required for both maintaining neuroepithelial cell identity and suppressing neuroblast formation. We further find that RPA or E2F1 inactivation in the neuroepithelial cells correlates with downregulation of Notch signaling activity, which appears to result from Numb mislocalization. Thus, we have shown that the transition from neuroepithelial cells to neuroblasts is directly regulated by cell cycle regulators and propose a model in which the inhibition of neuroepithelial cell cycle progression downregulates Notch signaling activity through Numb, which leads to the onset of neurogenesis.
Figures
Similar articles
-
Notch signaling regulates neuroepithelial stem cell maintenance and neuroblast formation in Drosophila optic lobe development.Dev Biol. 2011 Feb 15;350(2):414-28. doi: 10.1016/j.ydbio.2010.12.002. Epub 2010 Dec 10. Dev Biol. 2011. PMID: 21146517
-
Changes in Notch signaling coordinates maintenance and differentiation of the Drosophila larval optic lobe neuroepithelia.Dev Neurobiol. 2012 Nov;72(11):1376-90. doi: 10.1002/dneu.20995. Epub 2012 Jul 27. Dev Neurobiol. 2012. PMID: 22038743 Free PMC article.
-
Drosophila medulla neuroblast termination via apoptosis, differentiation, and gliogenic switch is scheduled by the depletion of the neuroepithelial stem cell pool.Elife. 2024 Jun 21;13:e96876. doi: 10.7554/eLife.96876. Elife. 2024. PMID: 38905123 Free PMC article.
-
Regulating the balance between symmetric and asymmetric stem cell division in the developing brain.Fly (Austin). 2011 Jul-Sep;5(3):237-41. doi: 10.4161/fly.5.3.15640. Epub 2011 Jul 1. Fly (Austin). 2011. PMID: 21502820 Review.
-
Cell cycle control of Notch signaling and the functional regionalization of the neuroepithelium during vertebrate neurogenesis.Int J Dev Biol. 2009;53(7):895-908. doi: 10.1387/ijdb.082721ml. Int J Dev Biol. 2009. PMID: 19598111 Review.
Cited by
-
Evidence for tissue-specific Jak/STAT target genes in Drosophila optic lobe development.Genetics. 2013 Dec;195(4):1291-306. doi: 10.1534/genetics.113.155945. Epub 2013 Sep 27. Genetics. 2013. PMID: 24077308 Free PMC article.
-
Broad Promotes Neuroepithelial Stem Cell Differentiation in the Drosophila Optic Lobe.Genetics. 2019 Nov;213(3):941-951. doi: 10.1534/genetics.119.302421. Epub 2019 Sep 17. Genetics. 2019. PMID: 31530575 Free PMC article.
-
Cell cycle networks link gene expression dysregulation, mutation, and brain maldevelopment in autistic toddlers.Mol Syst Biol. 2015 Dec 14;11(12):841. doi: 10.15252/msb.20156108. Mol Syst Biol. 2015. PMID: 26668231 Free PMC article.
-
Replication Protein A Presents Canonical Functions and Is Also Involved in the Differentiation Capacity of Trypanosoma cruzi.PLoS Negl Trop Dis. 2016 Dec 16;10(12):e0005181. doi: 10.1371/journal.pntd.0005181. eCollection 2016 Dec. PLoS Negl Trop Dis. 2016. PMID: 27984589 Free PMC article.
-
The Atr-Chek1 pathway inhibits axon regeneration in response to Piezo-dependent mechanosensation.Nat Commun. 2021 Jun 22;12(1):3845. doi: 10.1038/s41467-021-24131-7. Nat Commun. 2021. PMID: 34158506 Free PMC article.
References
-
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. - PubMed
-
- Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair. 2004;3:1015–1024. - PubMed
-
- Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36–42. - PubMed
-
- Brand AH, Livesey FJ. Neural stem cell biology in vertebrates and invertebrates: more alike than different? Neuron. 2011;70:719–729. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
