Reversal of mitochondrial proteomic loss in Type 1 diabetic heart with overexpression of phospholipid hydroperoxide glutathione peroxidase
- PMID: 23408027
- PMCID: PMC3627941
- DOI: 10.1152/ajpregu.00249.2012
Reversal of mitochondrial proteomic loss in Type 1 diabetic heart with overexpression of phospholipid hydroperoxide glutathione peroxidase
Abstract
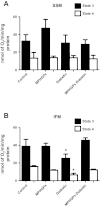
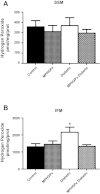
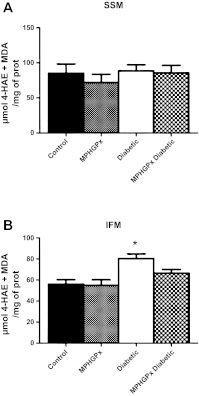
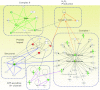
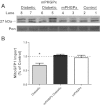
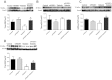
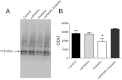
Mitochondrial dysfunction is a contributor to diabetic cardiomyopathy. Previously, we observed proteomic decrements within the inner mitochondrial membrane (IMM) and matrix of diabetic cardiac interfibrillar mitochondria (IFM) correlating with dysfunctional mitochondrial protein import. The goal of this study was to determine whether overexpression of mitochondria phospholipid hydroperoxide glutathione peroxidase 4 (mPHGPx), an antioxidant enzyme capable of scavenging membrane-associated lipid peroxides in the IMM, could reverse proteomic alterations, dysfunctional protein import, and ultimately, mitochondrial dysfunction associated with the diabetic heart. MPHGPx transgenic mice and controls were made diabetic by multiple low-dose streptozotocin injections and examined after 5 wk of hyperglycemia. Five weeks after hyperglycemia onset, in vivo analysis of cardiac contractile function revealed decreased ejection fraction and fractional shortening in diabetic hearts that was reversed with mPHGPx overexpression. MPHGPx overexpression increased electron transport chain function while attenuating hydrogen peroxide production and lipid peroxidation in diabetic mPHGPx IFM. MPHGPx overexpression lessened proteomic loss observed in diabetic IFM. Posttranslational modifications, including oxidations and deamidations, were attenuated in diabetic IFM with mPHGPx overexpression. Mitochondrial protein import dysfunction in diabetic IFM was reversed with mPHGPx overexpression correlating with protein import constituent preservation. Ingenuity Pathway Analyses indicated that oxidative phosphorylation, tricarboxylic acid cycle, and fatty acid oxidation processes most influenced in diabetic IFM were preserved by mPHGPx overexpression. Specific mitochondrial networks preserved included complex I and II, mitochondrial ultrastructure, and mitochondrial protein import. These results indicate that mPHGPx overexpression can preserve the mitochondrial proteome and provide cardioprotective benefits to the diabetic heart.
Figures

References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465, 1981 - PubMed
-
- Baseler WA, Dabkowski ER, Williamson CL, Croston TL, Thapa D, Powell MJ, Razunguzwa TT, Hollander JM. Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart: contribution of protein import dysfunction. Am J Physiol Regul Integr Comp Physiol 300: R186–R200, 2011 - PMC - PubMed
-
- Becker D, Krayl M, Strub A, Li Y, Mayer MP, Voos W. Impaired interdomain communication in mitochondrial Hsp70 results in the loss of inward-directed translocation force. J Biol Chem 284: 2934–2946, 2009 - PubMed
-
- Bohnert M, Rehling P, Guiard B, Herrmann JM, Pfanner N, van der Laan M. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr Biol 20: 1227–1232, 2010 - PubMed
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254, 1976 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

