Recognition of lipid A variants by the TLR4-MD-2 receptor complex
- PMID: 23408095
- PMCID: PMC3569842
- DOI: 10.3389/fcimb.2013.00003
Recognition of lipid A variants by the TLR4-MD-2 receptor complex
Abstract
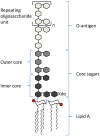
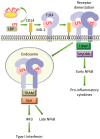
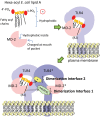
Lipopolysaccharide (LPS) is a component of the outer membrane of almost all Gram-negative bacteria and consists of lipid A, core sugars, and O-antigen. LPS is recognized by Toll-like receptor 4 (TLR4) and MD-2 on host innate immune cells and can signal to activate the transcription factor NFκB, leading to the production of pro-inflammatory cytokines that initiate and shape the adaptive immune response. Most of what is known about how LPS is recognized by the TLR4-MD-2 receptor complex on animal cells has been studied using Escherichia coli lipid A, which is a strong agonist of TLR4 signaling. Recent work from several groups, including our own, has shown that several important pathogenic bacteria can modify their LPS or lipid A molecules in ways that significantly alter TLR4 signaling to NFκB. Thus, it has been hypothesized that expression of lipid A variants is one mechanism by which pathogens modulate or evade the host immune response. Additionally, several key differences in the amino acid sequences of human and mouse TLR4-MD-2 receptors have been shown to alter the ability to recognize these variations in lipid A, suggesting a host-specific effect on the immune response to these pathogens. In this review, we provide an overview of lipid A variants from several human pathogens, how the basic structure of lipid A is recognized by mouse and human TLR4-MD-2 receptor complexes, as well as how alteration of this pattern affects its recognition by TLR4 and impacts the downstream immune response.
Keywords: LPS; TLR4; innate immunity; lipid A; signaling.
Figures

References
-
- Asensio C. J., Gaillard M. E., Moreno G., Bottero D., Zurita E., Rumbo M., et al. (2011). Outer membrane vesicles obtained from Bordetella pertussis Tohama expressing the lipid A deacylase PagL as a novel acellular vaccine candidate. Vaccine 29, 1649–1656 10.1016/j.vaccine.2010.12.068 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

