In silico Evaluation of Crosslinking Effects on Denaturant m(eq) values and ΔCp upon Protein Unfolding
- PMID: 23408172
- PMCID: PMC3558204
In silico Evaluation of Crosslinking Effects on Denaturant m(eq) values and ΔCp upon Protein Unfolding
Abstract
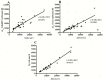
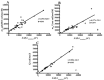
Important thermodynamic parameters including denaturant equilibrium m values (m(eq)) and heat capacity changes (ΔCp) can be predicted based on changes in Solvent Accessible Surface Area (SASA) upon unfolding. Crosslinks such as disulfide bonds influence the stability of the proteins by decreasing the entropy gain as well as reduction of SASA of unfolded state. The aim of the study was to develop mathematical models to predict the effect of crosslinks on ΔSASA and ultimately on m(eq) and ΔCp based on in silico methods. Changes of SASA upon computationally simulated unfolding were calculated for a set of 45 proteins with known m(eq) and ΔCp values and the effect of crosslinks on ΔSASA of unfolding was investigated. The results were used to predict the m(eq) of denaturation for guanidine hydrochloride and urea, as well as ΔCp for the studied proteins with overall error of 20%, 31% and 17%, respectively. The results of the current study were in close agreement with those obtained from the previous studies.
Keywords: Crosslinks; Disulfides; Protein stability; Thermodynamics.
Figures


Similar articles
-
Thermodynamic analysis of interactions between denaturants and protein surface exposed on unfolding: interpretation of urea and guanidinium chloride m-values and their correlation with changes in accessible surface area (ASA) using preferential interaction coefficients and the local-bulk domain model.Proteins. 2000;Suppl 4:72-85. doi: 10.1002/1097-0134(2000)41:4+<72::aid-prot70>3.0.co;2-7. Proteins. 2000. PMID: 11013402
-
Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding.Protein Sci. 1995 Oct;4(10):2138-48. doi: 10.1002/pro.5560041020. Protein Sci. 1995. PMID: 8535251 Free PMC article.
-
Linear free-energy model description of the conformational stability of uracil-DNA glycosylase inhibitor A thermodynamic characterization of interaction with denaturant and cold denaturation.Eur J Biochem. 1999 May;261(3):610-7. doi: 10.1046/j.1432-1327.1999.00271.x. Eur J Biochem. 1999. PMID: 10215876
-
Denaturant mediated unfolding of both native and molten globule states of maltose binding protein are accompanied by large deltaCp's.Protein Sci. 1999 Aug;8(8):1689-95. doi: 10.1110/ps.8.8.1689. Protein Sci. 1999. PMID: 10452613 Free PMC article.
-
A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states.Curr Protein Pept Sci. 2014;15(5):456-76. doi: 10.2174/1389203715666140327114232. Curr Protein Pept Sci. 2014. PMID: 24678666 Review.
References
-
- Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol Cell Proteomics. 2005;4(4):384–393. - PubMed
-
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6th ed. New York: W.H. Freeman; 2006.
-
- Morris MB, Dastmalchi S, Church WB. Rhodopsin: structure, signal transduction and oligomerisation. Int J Biochem Cell Biol. 2009;41(4):721–724. - PubMed
-
- Stefani M. Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta. 2004;1739(1):5–25. - PubMed
LinkOut - more resources
Full Text Sources
