The astonishing diversity of Ig classes and B cell repertoires in teleost fish
- PMID: 23408183
- PMCID: PMC3570791
- DOI: 10.3389/fimmu.2013.00028
The astonishing diversity of Ig classes and B cell repertoires in teleost fish
Abstract
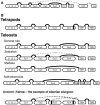
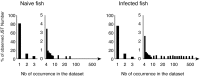
With lymphoid tissue anatomy different than mammals, and diverse adaptations to all aquatic environments, fish constitute a fascinating group of vertebrate to study the biology of B cell repertoires in a comparative perspective. Fish B lymphocytes express immunoglobulin (Ig) on their surface and secrete antigen-specific antibodies in response to immune challenges. Three antibody classes have been identified in fish, namely IgM, IgD, and IgT, while IgG, IgA, and IgE are absent. IgM and IgD have been found in all fish species analyzed, and thus seem to be primordial antibody classes. IgM and IgD are normally co-expressed from the same mRNA through alternative splicing, as in mammals. Tetrameric IgM is the main antibody class found in serum. Some species of fish also have IgT, which seems to exist only in fish and is specialized in mucosal immunity. IgM/IgD and IgT are expressed by two different sub-populations of B cells. The tools available to investigate B cell responses at the cellular level in fish are limited, but the progress of fish genomics has started to unravel a rich diversity of IgH and immunoglobulin light chain locus organization, which might be related to the succession of genome remodelings that occurred during fish evolution. Moreover, the development of deep sequencing techniques has allowed the investigation of the global features of the expressed fish B cell repertoires in zebrafish and rainbow trout, in steady state or after infection. This review provides a description of the organization of fish Ig loci, with a particular emphasis on their heterogeneity between species, and presents recent data on the structure of the expressed Ig repertoire in healthy and infected fish.
Keywords: B cells; antibody; evolution; fish; repertoire.
Figures


Similar articles
-
Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection.PLoS Pathog. 2013 Jan;9(1):e1003098. doi: 10.1371/journal.ppat.1003098. Epub 2013 Jan 10. PLoS Pathog. 2013. PMID: 23326228 Free PMC article.
-
Evolutionary and functional relationships of B cells from fish and mammals: insights into their novel roles in phagocytosis and presentation of particulate antigen.Infect Disord Drug Targets. 2012 Jun;12(3):200-12. doi: 10.2174/187152612800564419. Infect Disord Drug Targets. 2012. PMID: 22394174 Free PMC article. Review.
-
Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish.Proc Natl Acad Sci U S A. 2005 May 10;102(19):6919-24. doi: 10.1073/pnas.0500027102. Epub 2005 Apr 29. Proc Natl Acad Sci U S A. 2005. PMID: 15863615 Free PMC article.
-
Immunoglobulin (Ig) heavy chain gene locus and immune responses upon parasitic, bacterial and fungal infection in loach, Misgurnus anguillicaudatus.Fish Shellfish Immunol. 2019 Mar;86:1139-1150. doi: 10.1016/j.fsi.2018.12.064. Epub 2018 Dec 29. Fish Shellfish Immunol. 2019. PMID: 30599252
-
Immunoglobulins in teleosts.Immunogenetics. 2021 Feb;73(1):65-77. doi: 10.1007/s00251-020-01195-1. Epub 2021 Jan 13. Immunogenetics. 2021. PMID: 33439286 Review.
Cited by
-
Systematic characterization of immunoglobulin loci and deep sequencing of the expressed repertoire in the Atlantic cod (Gadus morhua).BMC Genomics. 2024 Jul 3;25(1):663. doi: 10.1186/s12864-024-10571-0. BMC Genomics. 2024. PMID: 38961347 Free PMC article.
-
Extreme genomic volatility characterizes the evolution of the immunoglobulin heavy chain locus in cyprinodontiform fishes.Proc Biol Sci. 2020 May 27;287(1927):20200489. doi: 10.1098/rspb.2020.0489. Epub 2020 May 13. Proc Biol Sci. 2020. PMID: 32396805 Free PMC article.
-
Fish Lymphocytes: An Evolutionary Equivalent of Mammalian Innate-Like Lymphocytes?Front Immunol. 2018 May 7;9:971. doi: 10.3389/fimmu.2018.00971. eCollection 2018. Front Immunol. 2018. PMID: 29867952 Free PMC article. Review.
-
To React or Not to React: The Dilemma of Fish Immune Systems Facing Myxozoan Infections.Front Immunol. 2021 Sep 16;12:734238. doi: 10.3389/fimmu.2021.734238. eCollection 2021. Front Immunol. 2021. PMID: 34603313 Free PMC article. Review.
-
IgM+ and IgT+ B Cell Traffic to the Heart during SAV Infection in Atlantic Salmon.Vaccines (Basel). 2020 Aug 31;8(3):493. doi: 10.3390/vaccines8030493. Vaccines (Basel). 2020. PMID: 32878234 Free PMC article.
References
-
- Abelli L., Picchietti S., Romano N., Mastrolia L., Scapigliati G. (1997). Immunohistochemistry of gut-associated lymphoid tissue of the sea bass Dicentrarchus labrax (L.). Fish Shellfish Immunol. 7, 235.10.1006/fsim.1996.0079 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

