Complete genome sequence of the sulfate-reducing firmicute Desulfotomaculum ruminis type strain (DL(T))
- PMID: 23408247
- PMCID: PMC3569383
- DOI: 10.4056/sigs.3226659
Complete genome sequence of the sulfate-reducing firmicute Desulfotomaculum ruminis type strain (DL(T))
Abstract
Desulfotomaculum ruminis Campbell and Postgate 1965 is a member of the large genus Desulfotomaculum which contains 30 species and is contained in the family Peptococcaceae. This species is of interest because it represents one of the few sulfate-reducing bacteria that have been isolated from the rumen. Here we describe the features of D. ruminis together with the complete genome sequence and annotation. The 3,969,014 bp long chromosome with a total of 3,901 protein-coding and 85 RNA genes is the second completed genome sequence of a type strain of the genus Desulfotomaculum to be published, and was sequenced as part of the DOE Joint Genome Institute Community Sequencing Program 2009.
Keywords: CSP 2009; Clostridiales; Peptococcaceae; anaerobic; hydrogen sulfide; incomplete oxidizer; mesophilic; mixotrophic; motile; sporulating; sulfate-reducer.
Figures
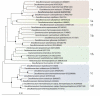

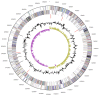


References
-
- Garrity G. NamesforLife. BrowserTool takes expertise out of the database and puts it right in the browser. Microbiol Today 2010; 37:9
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

