ErbB1-4-dependent EGF/neuregulin signals and their cross talk in the central nervous system: pathological implications in schizophrenia and Parkinson's disease
- PMID: 23408472
- PMCID: PMC3570895
- DOI: 10.3389/fncel.2013.00004
ErbB1-4-dependent EGF/neuregulin signals and their cross talk in the central nervous system: pathological implications in schizophrenia and Parkinson's disease
Abstract
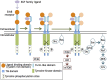
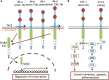
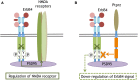
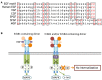
Ligands for ErbB1-4 receptor tyrosine kinases, such as epidermal growth factor (EGF) and neuregulins, regulate brain development and function. Thus, abnormalities in their signaling are implicated in the etiology or pathology of schizophrenia and Parkinson's disease. Among the ErbB receptors, ErbB1, and ErbB4 are expressed in dopamine and GABA neurons, while ErbB1, 2, and/or 3 are mainly present in oligodendrocytes, astrocytes, and their precursors. Thus, deficits in ErbB signaling might contribute to the neurological and psychiatric diseases stemming from these cell types. By incorporating the latest cancer molecular biology as well as our recent progress, we discuss signal cross talk between the ErbB1-4 subunits and their neurobiological functions in each cell type. The potential contribution of virus-derived cytokines (virokines) that mimic EGF and neuregulin-1 in brain diseases are also discussed.
Keywords: ErbB1-4; GABA; Parkinson's disease; dopamine; schizophrenia; virokine.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

