Myeloma is characterized by stage-specific alterations in DNA methylation that occur early during myelomagenesis
- PMID: 23408834
- PMCID: PMC4581585
- DOI: 10.4049/jimmunol.1202493
Myeloma is characterized by stage-specific alterations in DNA methylation that occur early during myelomagenesis
Abstract
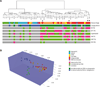
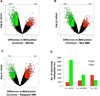
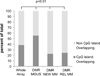
Epigenetic changes play important roles in carcinogenesis and influence initial steps in neoplastic transformation by altering genome stability and regulating gene expression. To characterize epigenomic changes during the transformation of normal plasma cells to myeloma, we modified the HpaII tiny fragment enrichment by ligation-mediated PCR assay to work with small numbers of purified primary marrow plasma cells. The nano-HpaII tiny fragment enrichment by ligation-mediated PCR assay was used to analyze the methylome of CD138(+) cells from 56 subjects representing premalignant (monoclonal gammopathy of uncertain significance), early, and advanced stages of myeloma, as well as healthy controls. Plasma cells from premalignant and early stages of myeloma were characterized by striking, widespread hypomethylation. Gene-specific hypermethylation was seen to occur in the advanced stages, and cell lines representative of relapsed cases were found to be sensitive to decitabine. Aberrant demethylation in monoclonal gammopathy of uncertain significance occurred primarily in CpG islands, whereas differentially methylated loci in cases of myeloma occurred predominantly outside of CpG islands and affected distinct sets of gene pathways, demonstrating qualitative epigenetic differences between premalignant and malignant stages. Examination of the methylation machinery revealed that the methyltransferase, DNMT3A, was aberrantly hypermethylated and underexpressed, but not mutated in myeloma. DNMT3A underexpression was also associated with adverse overall survival in a large cohort of patients, providing insights into genesis of hypomethylation in myeloma. These results demonstrate widespread, stage-specific epigenetic changes during myelomagenesis and suggest that early demethylation can be a potential contributor to genome instability seen in myeloma. We also identify DNMT3A expression as a novel prognostic biomarker and suggest that relapsed cases can be therapeutically targeted by hypomethylating agents.
Conflict of interest statement
The authors have no conflicts of interest to declare
Figures

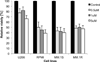
Similar articles
-
DNA methylation analysis determines the high frequency of genic hypomethylation and low frequency of hypermethylation events in plasma cell tumors.Cancer Res. 2010 Sep 1;70(17):6934-44. doi: 10.1158/0008-5472.CAN-10-0282. Epub 2010 Aug 24. Cancer Res. 2010. PMID: 20736376
-
Epigenetic silencing of miR-340-5p in multiple myeloma: mechanisms and prognostic impact.Clin Epigenetics. 2019 May 7;11(1):71. doi: 10.1186/s13148-019-0669-2. Clin Epigenetics. 2019. PMID: 31064412 Free PMC article.
-
Aberrant p15, p16, p53, and DAPK Gene Methylation in Myelomagenesis: Clinical and Prognostic Implications.Clin Lymphoma Myeloma Leuk. 2016 Dec;16(12):713-720.e2. doi: 10.1016/j.clml.2016.08.016. Epub 2016 Aug 10. Clin Lymphoma Myeloma Leuk. 2016. PMID: 27622827
-
Gene hypermethylation in multiple myeloma: lessons from a cancer pathway approach.Clin Lymphoma Myeloma. 2008 Dec;8(6):331-9. doi: 10.3816/CLM.2008.n.048. Clin Lymphoma Myeloma. 2008. PMID: 19064397 Review.
-
Decoding DNA methylation in epigenetics of multiple myeloma.Blood Rev. 2022 Jan;51:100872. doi: 10.1016/j.blre.2021.100872. Epub 2021 Jul 31. Blood Rev. 2022. PMID: 34384602 Review.
Cited by
-
Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers.Genome Res. 2015 Apr;25(4):478-87. doi: 10.1101/gr.180240.114. Epub 2015 Feb 2. Genome Res. 2015. PMID: 25644835 Free PMC article.
-
Clinicopathological significance of p15 promoter hypermethylation in multiple myeloma: a meta-analysis.Onco Targets Ther. 2016 Jul 1;9:4015-22. doi: 10.2147/OTT.S102733. eCollection 2016. Onco Targets Ther. 2016. PMID: 27445492 Free PMC article.
-
Physiological and pathological implications of 5-hydroxymethylcytosine in diseases.Oncotarget. 2016 Jul 26;7(30):48813-48831. doi: 10.18632/oncotarget.9281. Oncotarget. 2016. PMID: 27183914 Free PMC article. Review.
-
One Omics Approach Does Not Rule Them All: The Metabolome and the Epigenome Join Forces in Haematological Malignancies.Epigenomes. 2021 Oct 8;5(4):22. doi: 10.3390/epigenomes5040022. Epigenomes. 2021. PMID: 34968247 Free PMC article. Review.
-
MAGE-A inhibit apoptosis and promote proliferation in multiple myeloma through regulation of BIM and p21Cip1.Oncotarget. 2020 Feb 18;11(7):727-739. doi: 10.18632/oncotarget.27488. eCollection 2020 Feb 18. Oncotarget. 2020. PMID: 32133047 Free PMC article.
References
-
- Anguiano A, Tuchman SA, Acharya C, Salter K, Gasparetto C, Zhan F, Dhodapkar M, Nevins J, Barlogie B, Shaughnessy JD., Jr Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. J Clin Oncol. 2009;27:4197–4203. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

