Modulation of gene expression via overlapping binding sites exerted by ZNF143, Notch1 and THAP11
- PMID: 23408857
- PMCID: PMC3627581
- DOI: 10.1093/nar/gkt088
Modulation of gene expression via overlapping binding sites exerted by ZNF143, Notch1 and THAP11
Abstract
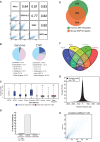
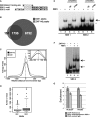
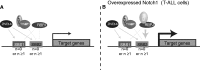
ZNF143 is a zinc-finger protein involved in the transcriptional regulation of both coding and non-coding genes from polymerase II and III promoters. Our study deciphers the genome-wide regulatory role of ZNF143 in relation with the two previously unrelated transcription factors Notch1/ICN1 and thanatos-associated protein 11 (THAP11) in several human and murine cells. We show that two distinct motifs, SBS1 and SBS2, are associated to ZNF143-binding events in promoters of >3000 genes. Without co-occupation, these sites are also bound by Notch1/ICN1 in T-lymphoblastic leukaemia cells as well as by THAP11, a factor involved in self-renewal of embryonic stem cells. We present evidence that ICN1 binding overlaps with ZNF143 binding events at the SBS1 and SBS2 motifs, whereas the overlap occurs only at SBS2 for THAP11. We demonstrate that the three factors modulate expression of common target genes through the mutually exclusive occupation of overlapping binding sites. The model we propose predicts that the binding competition between the three factors controls biological processes such as rapid cell growth of both neoplastic and stem cells. Overall, our study establishes a novel relationship between ZNF143, THAP11 and ICN1 and reveals important insights into ZNF143-mediated gene regulation.
Figures




Similar articles
-
ZNF143 in Chromatin Looping and Gene Regulation.Front Genet. 2020 Apr 7;11:338. doi: 10.3389/fgene.2020.00338. eCollection 2020. Front Genet. 2020. PMID: 32318100 Free PMC article. Review.
-
Genomic Determinants of THAP11/ZNF143/HCFC1 Complex Recruitment to Chromatin.Mol Cell Biol. 2015 Dec;35(24):4135-46. doi: 10.1128/MCB.00477-15. Epub 2015 Sep 28. Mol Cell Biol. 2015. PMID: 26416877 Free PMC article.
-
Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells.Proc Natl Acad Sci U S A. 2011 Sep 6;108(36):14908-13. doi: 10.1073/pnas.1109023108. Epub 2011 Jul 7. Proc Natl Acad Sci U S A. 2011. PMID: 21737748 Free PMC article.
-
Biochemical and cellular characterization of transcription factors binding to the hyperconserved core promoter-associated M4 motif.BMC Genomics. 2016 Aug 30;17(1):693. doi: 10.1186/s12864-016-3033-3. BMC Genomics. 2016. PMID: 27576892 Free PMC article.
-
The ubiquitous transcriptional protein ZNF143 activates a diversity of genes while assisting to organize chromatin structure.Gene. 2021 Feb 15;769:145205. doi: 10.1016/j.gene.2020.145205. Epub 2020 Oct 5. Gene. 2021. PMID: 33031894 Review.
Cited by
-
Systematic identification of gene family regulators in mouse and human embryonic stem cells.Nucleic Acids Res. 2016 May 19;44(9):4080-9. doi: 10.1093/nar/gkw259. Epub 2016 Apr 15. Nucleic Acids Res. 2016. PMID: 27084933 Free PMC article.
-
Tissue-Specific Actions of Pax6 on Proliferation and Differentiation Balance in Developing Forebrain Are Foxg1 Dependent.iScience. 2018 Dec 21;10:171-191. doi: 10.1016/j.isci.2018.11.031. Epub 2018 Nov 22. iScience. 2018. PMID: 30529950 Free PMC article.
-
Putative looping factor ZNF143/ZFP143 is an essential transcriptional regulator with no looping function.Mol Cell. 2025 Jan 2;85(1):9-23.e9. doi: 10.1016/j.molcel.2024.11.032. Epub 2024 Dec 20. Mol Cell. 2025. PMID: 39708803 Free PMC article.
-
ZNF143 in Chromatin Looping and Gene Regulation.Front Genet. 2020 Apr 7;11:338. doi: 10.3389/fgene.2020.00338. eCollection 2020. Front Genet. 2020. PMID: 32318100 Free PMC article. Review.
-
Hi-TrAC reveals division of labor of transcription factors in organizing chromatin loops.Nat Commun. 2022 Nov 5;13(1):6679. doi: 10.1038/s41467-022-34276-8. Nat Commun. 2022. PMID: 36335136 Free PMC article.
References
-
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. - PubMed
-
- Lenhard B, Sandelin A, Carninci P. Regulatory elements: Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13:233–245. - PubMed
-
- Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

