Linking proteomic and transcriptional data through the interactome and epigenome reveals a map of oncogene-induced signaling
- PMID: 23408876
- PMCID: PMC3567149
- DOI: 10.1371/journal.pcbi.1002887
Linking proteomic and transcriptional data through the interactome and epigenome reveals a map of oncogene-induced signaling
Abstract
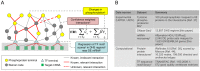
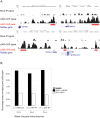
Cellular signal transduction generally involves cascades of post-translational protein modifications that rapidly catalyze changes in protein-DNA interactions and gene expression. High-throughput measurements are improving our ability to study each of these stages individually, but do not capture the connections between them. Here we present an approach for building a network of physical links among these data that can be used to prioritize targets for pharmacological intervention. Our method recovers the critical missing links between proteomic and transcriptional data by relating changes in chromatin accessibility to changes in expression and then uses these links to connect proteomic and transcriptome data. We applied our approach to integrate epigenomic, phosphoproteomic and transcriptome changes induced by the variant III mutation of the epidermal growth factor receptor (EGFRvIII) in a cell line model of glioblastoma multiforme (GBM). To test the relevance of the network, we used small molecules to target highly connected nodes implicated by the network model that were not detected by the experimental data in isolation and we found that a large fraction of these agents alter cell viability. Among these are two compounds, ICG-001, targeting CREB binding protein (CREBBP), and PKF118-310, targeting β-catenin (CTNNB1), which have not been tested previously for effectiveness against GBM. At the level of transcriptional regulation, we used chromatin immunoprecipitation sequencing (ChIP-Seq) to experimentally determine the genome-wide binding locations of p300, a transcriptional co-regulator highly connected in the network. Analysis of p300 target genes suggested its role in tumorigenesis. We propose that this general method, in which experimental measurements are used as constraints for building regulatory networks from the interactome while taking into account noise and missing data, should be applicable to a wide range of high-throughput datasets.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Gil JM, Rego AC (2008) Mechanisms of neurodegeneration in Huntington's disease. Eur J Neurosci 27: 2803–2820 Available:http://dx.doi.org/10.1111/j.1460-9568.2008.06310.x. - DOI - PubMed
-
- Imarisio S, Carmichael J, Korolchuk V, Chen C-W, Saiki S, et al. (2008) Huntington's disease: from pathology and genetics to potential therapies. Biochem J 412: 191–209 Available:http://dx.doi.org/10.1042/BJ20071619. - DOI - PubMed
-
- Schinner S, Scherbaum WA, Bornstein SR, Barthel A (2005) Molecular mechanisms of insulin resistance. Diabet Med 22: 674–682 Available:http://dx.doi.org/10.1111/j.1464-5491.2005.01566.x. - DOI - PubMed
-
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 Available:http://dx.doi.org/10.1016/j.cell.2011.02.013. - DOI - PubMed
-
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

