Structural and functional analysis of the CspB protease required for Clostridium spore germination
- PMID: 23408892
- PMCID: PMC3567191
- DOI: 10.1371/journal.ppat.1003165
Structural and functional analysis of the CspB protease required for Clostridium spore germination
Abstract
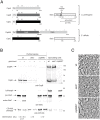
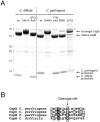
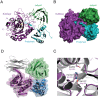
Spores are the major transmissive form of the nosocomial pathogen Clostridium difficile, a leading cause of healthcare-associated diarrhea worldwide. Successful transmission of C. difficile requires that its hardy, resistant spores germinate into vegetative cells in the gastrointestinal tract. A critical step during this process is the degradation of the spore cortex, a thick layer of peptidoglycan surrounding the spore core. In Clostridium sp., cortex degradation depends on the proteolytic activation of the cortex hydrolase, SleC. Previous studies have implicated Csps as being necessary for SleC cleavage during germination; however, their mechanism of action has remained poorly characterized. In this study, we demonstrate that CspB is a subtilisin-like serine protease whose activity is essential for efficient SleC cleavage and C. difficile spore germination. By solving the first crystal structure of a Csp family member, CspB, to 1.6 Å, we identify key structural domains within CspB. In contrast with all previously solved structures of prokaryotic subtilases, the CspB prodomain remains tightly bound to the wildtype subtilase domain and sterically occludes a catalytically competent active site. The structure, combined with biochemical and genetic analyses, reveals that Csp proteases contain a unique jellyroll domain insertion critical for stabilizing the protease in vitro and in C. difficile. Collectively, our study provides the first molecular insight into CspB activity and function. These studies may inform the development of inhibitors that can prevent clostridial spore germination and thus disease transmission.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Carroll KC, Bartlett JG (2011) Biology of Clostridium difficile: implications for epidemiology and diagnosis. Annu Rev Microbiol 65: 501–521. - PubMed
-
- Kelly CP, LaMont JT (2008) Clostridium difficile–more difficult than ever. N Engl J Med 359: 1932–1940. - PubMed
-
- Rupnik M, Wilcox MH, Gerding DN (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7: 526–536. - PubMed
-
- Ghantoji SS, Sail K, Lairson DR, DuPont HL, Garey KW (2010) Economic healthcare costs of Clostridium difficile infection: a systematic review. J Hosp Infect 74: 309–318. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

