The Mub1/Ubr2 ubiquitin ligase complex regulates the conserved Dsn1 kinetochore protein
- PMID: 23408894
- PMCID: PMC3567142
- DOI: 10.1371/journal.pgen.1003216
The Mub1/Ubr2 ubiquitin ligase complex regulates the conserved Dsn1 kinetochore protein
Abstract
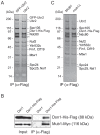
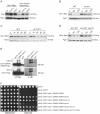
The kinetochore is the macromolecular complex that assembles onto centromeric DNA and orchestrates the segregation of duplicated chromosomes. More than 60 components make up the budding yeast kinetochore, including inner kinetochore proteins that bind to centromeric chromatin and outer proteins that directly interact with microtubules. However, little is known about how these components assemble into a functional kinetochore and whether there are quality control mechanisms that monitor kinetochore integrity. We previously developed a method to isolate kinetochore particles via purification of the conserved Dsn1 kinetochore protein. We find that the Mub1/Ubr2 ubiquitin ligase complex associates with kinetochore particles through the CENP-C(Mif2) protein. Although Mub1/Ubr2 are not stable kinetochore components in vivo, they regulate the levels of the conserved outer kinetochore protein Dsn1 via ubiquitylation. Strikingly, a deletion of Mub1/Ubr2 restores the levels and viability of a mutant Dsn1 protein, reminiscent of quality control systems that target aberrant proteins for degradation. Consistent with this, Mub1/Ubr2 help to maintain viability when kinetochores are defective. Together, our data identify a previously unknown regulatory mechanism for the conserved Dsn1 kinetochore protein. We propose that Mub1/Ubr2 are part of a quality control system that monitors kinetochore integrity, thus ensuring genomic stability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

