Drosophila Yemanuclein and HIRA cooperate for de novo assembly of H3.3-containing nucleosomes in the male pronucleus
- PMID: 23408912
- PMCID: PMC3567178
- DOI: 10.1371/journal.pgen.1003285
Drosophila Yemanuclein and HIRA cooperate for de novo assembly of H3.3-containing nucleosomes in the male pronucleus
Abstract
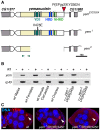
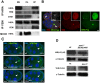
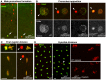
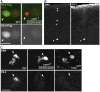
The differentiation of post-meiotic spermatids in animals is characterized by a unique reorganization of their nuclear architecture and chromatin composition. In many species, the formation of sperm nuclei involves the massive replacement of nucleosomes with protamines, followed by a phase of extreme nuclear compaction. At fertilization, the reconstitution of a nucleosome-based paternal chromatin after the removal of protamines requires the deposition of maternally provided histones before the first round of DNA replication. This process exclusively uses the histone H3 variant H3.3 and constitutes a unique case of genome-wide replication-independent (RI) de novo chromatin assembly. We had previously shown that the histone H3.3 chaperone HIRA plays a central role for paternal chromatin assembly in Drosophila. Although several conserved HIRA-interacting proteins have been identified from yeast to human, their conservation in Drosophila, as well as their actual implication in this highly peculiar RI nucleosome assembly process, is an open question. Here, we show that Yemanuclein (YEM), the Drosophila member of the Hpc2/Ubinuclein family, is essential for histone deposition in the male pronucleus. yem loss of function alleles affect male pronucleus formation in a way remarkably similar to Hira mutants and abolish RI paternal chromatin assembly. In addition, we demonstrate that HIRA and YEM proteins interact and are mutually dependent for their targeting to the decondensing male pronucleus. Finally, we show that the alternative ATRX/XNP-dependent H3.3 deposition pathway is not involved in paternal chromatin assembly, thus underlining the specific implication of the HIRA/YEM complex for this essential step of zygote formation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization.PLoS Genet. 2007 Oct;3(10):1991-2006. doi: 10.1371/journal.pgen.0030182. Epub 2007 Sep 10. PLoS Genet. 2007. PMID: 17967064 Free PMC article.
-
ASF1 is required to load histones on the HIRA complex in preparation of paternal chromatin assembly at fertilization.Epigenetics Chromatin. 2018 May 11;11(1):19. doi: 10.1186/s13072-018-0189-x. Epigenetics Chromatin. 2018. PMID: 29751847 Free PMC article.
-
The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus.Nature. 2005 Oct 27;437(7063):1386-90. doi: 10.1038/nature04059. Nature. 2005. PMID: 16251970
-
A Molecular Prospective for HIRA Complex Assembly and H3.3-Specific Histone Chaperone Function.J Mol Biol. 2017 Jun 30;429(13):1924-1933. doi: 10.1016/j.jmb.2016.11.010. Epub 2016 Nov 19. J Mol Biol. 2017. PMID: 27871933 Free PMC article. Review.
-
Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly.Cell Mol Life Sci. 2008 Feb;65(3):414-44. doi: 10.1007/s00018-007-7305-6. Cell Mol Life Sci. 2008. PMID: 17955179 Free PMC article. Review.
Cited by
-
Single Nucleotide Polymorphisms in the HIRA Gene Affect Litter Size in Small Tail Han Sheep.Animals (Basel). 2018 May 4;8(5):71. doi: 10.3390/ani8050071. Animals (Basel). 2018. PMID: 29734691 Free PMC article.
-
Multiple Roles of dXNP and dADD1-Drosophila Orthologs of ATRX Chromatin Remodeler.Int J Mol Sci. 2023 Nov 18;24(22):16486. doi: 10.3390/ijms242216486. Int J Mol Sci. 2023. PMID: 38003676 Free PMC article. Review.
-
dBRWD3 Regulates Tissue Overgrowth and Ectopic Gene Expression Caused by Polycomb Group Mutations.PLoS Genet. 2016 Sep 2;12(9):e1006262. doi: 10.1371/journal.pgen.1006262. eCollection 2016 Sep. PLoS Genet. 2016. PMID: 27588417 Free PMC article.
-
Reduced histone gene copy number disrupts Drosophila Polycomb function.bioRxiv [Preprint]. 2023 Mar 28:2023.03.28.534544. doi: 10.1101/2023.03.28.534544. bioRxiv. 2023. Update in: Genetics. 2023 Aug 9;224(4):iyad106. doi: 10.1093/genetics/iyad106. PMID: 37034607 Free PMC article. Updated. Preprint.
-
Reduced histone gene copy number disrupts Drosophila Polycomb function.Genetics. 2023 Aug 9;224(4):iyad106. doi: 10.1093/genetics/iyad106. Genetics. 2023. PMID: 37279945 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

