Multi-species comparative analysis of the equine ACE gene identifies a highly conserved potential transcription factor binding site in intron 16
- PMID: 23408978
- PMCID: PMC3568152
- DOI: 10.1371/journal.pone.0055434
Multi-species comparative analysis of the equine ACE gene identifies a highly conserved potential transcription factor binding site in intron 16
Abstract
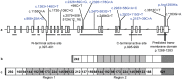
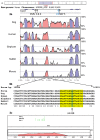
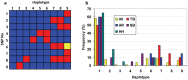
Angiotensin converting enzyme (ACE) is essential for control of blood pressure. The human ACE gene contains an intronic Alu indel (I/D) polymorphism that has been associated with variation in serum enzyme levels, although the functional mechanism has not been identified. The polymorphism has also been associated with cardiovascular disease, type II diabetes, renal disease and elite athleticism. We have characterized the ACE gene in horses of breeds selected for differing physical abilities. The equine gene has a similar structure to that of all known mammalian ACE genes. Nine common single nucleotide polymorphisms (SNPs) discovered in pooled DNA were found to be inherited in nine haplotypes. Three of these SNPs were located in intron 16, homologous to that containing the Alu polymorphism in the human. A highly conserved 18 bp sequence, also within that intron, was identified as being a potential binding site for the transcription factors Oct-1, HFH-1 and HNF-3β, and lies within a larger area of higher than normal homology. This putative regulatory element may contribute to regulation of the documented inter-individual variation in human circulating enzyme levels, for which a functional mechanism is yet to be defined. Two equine SNPs occurred within the conserved area in intron 16, although neither of them disrupted the putative binding site. We propose a possible regulatory mechanism of the ACE gene in mammalian species which was previously unknown. This advance will allow further analysis leading to a better understanding of the mechanisms underpinning the associations seen between the human Alu polymorphism and enzyme levels, cardiovascular disease states and elite athleticism.
Conflict of interest statement
Figures
Similar articles
-
Exonization of AluYa5 in the human ACE gene requires mutations in both 3' and 5' splice sites and is facilitated by a conserved splicing enhancer.Nucleic Acids Res. 2005 Jul 14;33(12):3897-906. doi: 10.1093/nar/gki707. Print 2005. Nucleic Acids Res. 2005. PMID: 16027113 Free PMC article.
-
Novel polymorphisms in UTR and coding region of inducible heat shock protein 70.1 gene in tropically adapted Indian zebu cattle (Bos indicus) and riverine buffalo (Bubalus bubalis).Gene. 2013 Sep 25;527(2):606-15. doi: 10.1016/j.gene.2013.05.078. Epub 2013 Jun 19. Gene. 2013. PMID: 23792016
-
Sequence variation in the human angiotensin converting enzyme.Nat Genet. 1999 May;22(1):59-62. doi: 10.1038/8760. Nat Genet. 1999. PMID: 10319862
-
Angiotensin converting enzyme gene insertion/deletion polymorphism and cardiovascular disease: therapeutic implications.Drugs. 2002;62(7):977-93. doi: 10.2165/00003495-200262070-00001. Drugs. 2002. PMID: 11985486 Review.
-
New aspects on angiotensin-converting enzyme: from gene to disease.Clin Chem Lab Med. 2002 Mar;40(3):256-65. doi: 10.1515/CCLM.2002.042. Clin Chem Lab Med. 2002. PMID: 12005216 Review.
Cited by
-
Functionally conserved enhancers with divergent sequences in distant vertebrates.BMC Genomics. 2015 Oct 30;16:882. doi: 10.1186/s12864-015-2070-7. BMC Genomics. 2015. PMID: 26519295 Free PMC article.
-
An integrative computational approach for prioritization of genomic variants.PLoS One. 2014 Dec 15;9(12):e114903. doi: 10.1371/journal.pone.0114903. eCollection 2014. PLoS One. 2014. PMID: 25506935 Free PMC article.
References
-
- Cole J, Ertoy D, Bernstein KE (2000) Insights derived from ACE knockout mice. J Renin Angiotensin Aldostrone Syst 1: 137–141. - PubMed
-
- Cambien F, Costerousse O, Tiret L, Poirier O, Lecerf L, et al. (1994) Plasma level and gene polymorphism of angiotensin-converting enzyme in relation to myocardial infarction. Circulation 90: 669–676. - PubMed
-
- Cambien F, Poirier O, Lecerf L, Evans A, Cambou J, et al. (1992) Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature 359: 641–644. - PubMed
-
- Evans A, Poirier O, Kee F, Lecerf L, McCrum E, et al. (1994) Polymorphisms of the angiotensin-converting enzyme gene in subjects who die from coronary heart disease. Q J Med 87: 211–214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

