Quantitative 1H-NMR-metabolomics reveals extensive metabolic reprogramming and the effect of the aquaglyceroporin FPS1 in ethanol-stressed yeast cells
- PMID: 23408980
- PMCID: PMC3568136
- DOI: 10.1371/journal.pone.0055439
Quantitative 1H-NMR-metabolomics reveals extensive metabolic reprogramming and the effect of the aquaglyceroporin FPS1 in ethanol-stressed yeast cells
Abstract
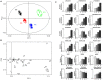
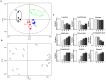
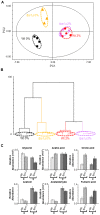
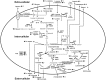
A metabolomic analysis using high resolution 1H NMR spectroscopy coupled with multivariate statistical analysis was used to characterize the alterations in the endo- and exo-metabolome of S. cerevisiae BY4741 during the exponential phase of growth in minimal medium supplemented with different ethanol concentrations (0, 2, 4 and 6% v/v). This study provides evidence that supports the notion that ethanol stress induces reductive stress in yeast cells, which, in turn, appears to be counteracted by the increase in the rate of NAD+ regenerating bioreactions. Metabolomics data also shows increased intra- and extra-cellular accumulation of most amino acids and TCA cycle intermediates in yeast cells growing under ethanol stress suggesting a state of overflow metabolism in turn of the pyruvate branch-point. Given its previous implication in ethanol stress resistance in yeast, this study also focused on the effect of the expression of the aquaglyceroporin encoded by FPS1 in the yeast metabolome, in the absence or presence of ethanol stress. The metabolomics data collected herein shows that the deletion of the FPS1 gene in the absence of ethanol stress partially mimics the effect of ethanol stress in the parental strain. Moreover, the results obtained suggest that the reported action of Fps1 in mediating the passive diffusion of glycerol is a key factor in the maintenance of redox balance, an important feature for ethanol stress resistance, and may interfere with the ability of the yeast cell to accumulate trehalose. Overall, the obtained results corroborate the idea that metabolomic approaches may be crucial tools to understand the function and/or the effect of membrane transporters/porins, such as Fps1, and may be an important tool for the clear-cut design of improved process conditions and more robust yeast strains aiming to optimize industrial fermentation performance.
Conflict of interest statement
Figures


References
-
- Gibson BR, Lawrence SJ, Leclaire JP, Powell CD, Smart KA (2007) Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiology Reviews 31: 535–569. - PubMed
-
- Stanley D, Chambers PJ, Stanley GA, Borneman A, Fraser S (2010) Transcriptional changes associated with ethanol tolerance in Saccharomyces cerevisiae . Applied Microbiology and Biotechnology 88: 231–239. - PubMed
-
- van Uden N (1985) Ethanol toxicity and ethanol tolerance in yeasts. Annual Reports on Fermentation Processes 8: 11–58.
-
- Gawel R, Francis L, Waters EJ (2007) Statistical correlations between the in-mouth textural characteristics and the chemical composition of Shiraz wines. Journal of Agriculture Food and Chemistry 55: 2683–2687. - PubMed
-
- Gawel R, van Sluyter S, Waters EJ (2007) The effects of ethanol and glycerol on the body and other sensory characteristics of Riesling wines. Australian Journal of Grape and Wine Research 13: 38–45.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

