Alterations in the colonic microbiota in response to osmotic diarrhea
- PMID: 23409050
- PMCID: PMC3568139
- DOI: 10.1371/journal.pone.0055817
Alterations in the colonic microbiota in response to osmotic diarrhea
Abstract
Background & aims: Diseases of the human gastrointestinal (GI) tract are often accompanied by diarrhea with profound alterations in the GI microbiota termed dysbiosis. Whether dysbiosis is due to the disease itself or to the accompanying diarrhea remains elusive. With this study we characterized the net effects of osmotic diarrhea on the composition of the GI microbiota in the absence of disease.
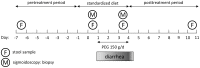
Methods: We induced osmotic diarrhea in four healthy adults by oral administration of polyethylene glycol 4000 (PEG). Stool as well as mucosa specimens were collected before, during and after diarrhea and 16S rDNA-based microbial community profiling was used to assess the microbial community structure.
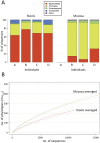
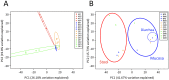
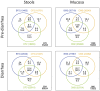
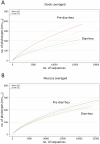
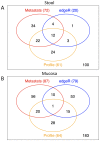
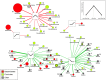
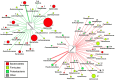
Results: Stool and mucosal microbiotas were strikingly different, with Firmicutes dominating the mucosa and Bacteroidetes the stools. Osmotic diarrhea decreased phylotype richness and showed a strong tendency to equalize the otherwise individualized microbiotas on the mucosa. Moreover, diarrhea led to significant relative shifts in the phyla Bacteroidetes and Firmicutes and to a relative increase in the abundance of Proteobacteria on the mucosa, a phenomenon also noted in several inflammatory and diarrheal GI diseases.
Conclusions: Changes in microbial community structure induced by osmotic diarrhea are profound and show similarities to changes observed in other GI diseases including IBD. These effects so must be considered when specimens from diarrheal diseases (i.e. obtained by stratification of samples according to diarrheal status) or conditions wherein bowel preparations like PEG (i.e. specimens obtained during endoscopy) are used.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

