Characterization of Capsicum annuum genetic diversity and population structure based on parallel polymorphism discovery with a 30K unigene Pepper GeneChip
- PMID: 23409153
- PMCID: PMC3568043
- DOI: 10.1371/journal.pone.0056200
Characterization of Capsicum annuum genetic diversity and population structure based on parallel polymorphism discovery with a 30K unigene Pepper GeneChip
Abstract
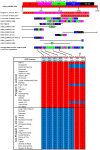
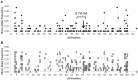
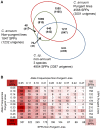
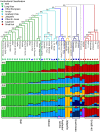
The widely cultivated pepper, Capsicum spp., important as a vegetable and spice crop world-wide, is one of the most diverse crops. To enhance breeding programs, a detailed characterization of Capsicum diversity including morphological, geographical and molecular data is required. Currently, molecular data characterizing Capsicum genetic diversity is limited. The development and application of high-throughput genome-wide markers in Capsicum will facilitate more detailed molecular characterization of germplasm collections, genetic relationships, and the generation of ultra-high density maps. We have developed the Pepper GeneChip® array from Affymetrix for polymorphism detection and expression analysis in Capsicum. Probes on the array were designed from 30,815 unigenes assembled from expressed sequence tags (ESTs). Our array design provides a maximum redundancy of 13 probes per base pair position allowing integration of multiple hybridization values per position to detect single position polymorphism (SPP). Hybridization of genomic DNA from 40 diverse C. annuum lines, used in breeding and research programs, and a representative from three additional cultivated species (C. frutescens, C. chinense and C. pubescens) detected 33,401 SPP markers within 13,323 unigenes. Among the C. annuum lines, 6,426 SPPs covering 3,818 unigenes were identified. An estimated three-fold reduction in diversity was detected in non-pungent compared with pungent lines, however, we were able to detect 251 highly informative markers across these C. annuum lines. In addition, an 8.7 cM region without polymorphism was detected around Pun1 in non-pungent C. annuum. An analysis of genetic relatedness and diversity using the software Structure revealed clustering of the germplasm which was confirmed with statistical support by principle components analysis (PCA) and phylogenetic analysis. This research demonstrates the effectiveness of parallel high-throughput discovery and application of genome-wide transcript-based markers to assess genetic and genomic features among Capsicum annuum.
Conflict of interest statement
Figures

References
-
- Bosland PW, Votava E (2000) Peppers: Vegetable and spice capsicums. Oxford, Wallingford: Cabi.
-
- Perry L, Dickau R, Zarrillo S, Holst I, Pearsall DM, et al. (2007) Starch fossils and the domestication and dispersal of chili peppers (Capsicum spp. L.) in the americas. Science 315: 986–988. - PubMed
-
- Eshbaugh WH (1993) Peppers: History and exploitation of a serendipitous new crop discovery. In: Janick J, Simon JE, editors. New crops. New York, USA: Wiley pp. 132–139.
-
- Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, et al. (2008) A molecular phylogeny of the solanaceae. Taxon 57: 1159–1181.
-
- USDA-ARS (2011) Grin species records of Capsicum. Beltsville, Maryland: National Germplasm Resources Laboratory.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

