Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia
- PMID: 23410974
- PMCID: PMC3597385
- DOI: 10.1016/j.ccr.2013.01.011
Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia
Abstract
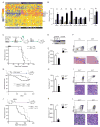
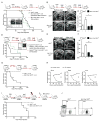
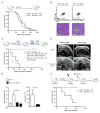
Most patients with acute lymphoblastic leukemia (ALL) fail current treatments highlighting the need for better therapies. Because oncogenic signaling activates a p53-dependent DNA damage response and apoptosis, leukemic cells must devise appropriate countermeasures. We show here that growth factor independence 1 (Gfi1) can serve such a function because Gfi1 ablation exacerbates p53 responses and lowers the threshold for p53-induced cell death. Specifically, Gfi1 restricts p53 activity and expression of proapoptotic p53 targets such as Bax, Noxa (Pmaip1), and Puma (Bbc3). Subsequently, Gfi1 ablation cures mice from leukemia and limits the expansion of primary human T-ALL xenografts in mice. This suggests that targeting Gfi1 could improve the prognosis of patients with T-ALL or other lymphoid leukemias.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. - PubMed
-
- Bartkova J, Horejsi Z, Sehested M, Nesland JM, Rajpert-De Meyts E, Skakkebaek NE, Stucki M, Jackson S, Lukas J, Bartek J. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene. 2007;26:7414–7422. - PubMed
-
- Blyth K, Terry A, Mackay N, Vaillant F, Bell M, Cameron ER, Neil JC, Stewart M. Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene. 2001;20:295–302. - PubMed
-
- Chakraborty J, Okonta H, Bagalb H, Lee SJ, Fink B, Changanamkandat R, Duggan J. Retroviral gene insertion in breast milk mediated lymphomagenesis. Virology. 2008;377:100–109. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 CA159845/CA/NCI NIH HHS/United States
- 079249/WT_/Wellcome Trust/United Kingdom
- ZIA AI001169/ImNIH/Intramural NIH HHS/United States
- G0900729/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- T32 CA117846/CA/NCI NIH HHS/United States
- 12765/CRUK_/Cancer Research UK/United Kingdom
- CA105152/CA/NCI NIH HHS/United States
- MOP-111011/CAPMC/ CIHR/Canada
- R01 CA105152/CA/NCI NIH HHS/United States
- P30 DK090971/DK/NIDDK NIH HHS/United States
- MC_PC_12009/MRC_/Medical Research Council/United Kingdom
- T32-CA117846/CA/NCI NIH HHS/United States
- CA159845/CA/NCI NIH HHS/United States
- 100140/WT_/Wellcome Trust/United Kingdom
- MOP-84238/CAPMC/ CIHR/Canada
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

