In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy
- PMID: 23413883
- PMCID: PMC3925062
- DOI: 10.1021/pr3011638
In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy
Abstract
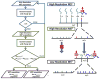
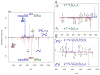
Protein interaction topologies are critical determinants of biological function. Large-scale or proteome-wide measurements of protein interaction topologies in cells currently pose an unmet challenge that could dramatically improve understanding of complex biological systems. A primary impediment includes direct protein topology and interaction measurements from living systems since interactions that lack biological significance may be introduced during cell lysis. Furthermore, many biologically relevant protein interactions will likely not survive the lysis/sample preparation and may only be measured with in vivo methods. As a step toward meeting this challenge, a new mass spectrometry method called Real-time Analysis for Cross-linked peptide Technology (ReACT) has been developed that enables assignment of cross-linked peptides "on-the-fly". Using ReACT, 708 unique cross-linked (<5% FDR) peptide pairs were identified from cross-linked E. coli cells. These data allow assembly of the first protein interaction network that also contains topological features of every interaction, as it existed in cells during cross-linker application. Of the identified interprotein cross-linked peptide pairs, 40% are derived from known interactions and provide new topological data that can help visualize how these interactions exist in cells. Other identified cross-linked peptide pairs are from proteins known to be involved within the same complex, but yield newly discovered direct physical interactors. ReACT enables the first view of these interactions inside cells, and the results acquired with this method suggest cross-linking can play a major role in future efforts to map the interactome in cells.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Ellis RJ. Macromolecular crowding: obvious but under-appreciated. Trends Biochem Sci. 2001;26(10):597–604. - PubMed
-
- Ikeya T, Sasaki A, Sakakibara D, Shigemitsu Y, Hamatsu J, Hanashima T, Mishima M, Yoshimasu M, Hayashi N, Mikawa T, Nietlispach D, Walchli M, Smith BO, Shirakawa M, Guntert P, Ito Y. NMR protein structure determination in living E. coli cells using nonlinear sampling. Nat Protoc. 2010;5(6):1051–60. - PubMed
-
- Robinson KE, Reardon PN, Spicer LD. In-cell NMR spectroscopy in Escherichia coli. Methods Mol Biol. 2012;831:261–77. - PubMed
-
- Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447(7147):1021–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

