Signal recognition particle: an essential protein-targeting machine
- PMID: 23414305
- PMCID: PMC3805129
- DOI: 10.1146/annurev-biochem-072711-164732
Signal recognition particle: an essential protein-targeting machine
Abstract
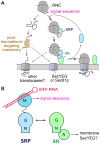
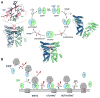
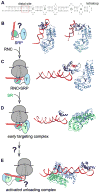
The signal recognition particle (SRP) and its receptor compose a universally conserved and essential cellular machinery that couples the synthesis of nascent proteins to their proper membrane localization. The past decade has witnessed an explosion in in-depth mechanistic investigations of this targeting machine at increasingly higher resolutions. In this review, we summarize recent work that elucidates how the SRP and SRP receptor interact with the cargo protein and the target membrane, respectively, and how these interactions are coupled to a novel GTPase cycle in the SRP·SRP receptor complex to provide the driving force and enhance the fidelity of this fundamental cellular pathway. We also discuss emerging frontiers in which important questions remain to be addressed.
Figures
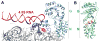


References
-
- Cross BCS, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nature Rev Mol Cell Biol. 2009;10:255–64. - PubMed
-
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–67. - PubMed
-
- Pool MR. Signal recognition particles in chloroplasts, bacteria, yeast and mammals. Mole Membr Biol. 2005;22:3–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

