A human skeletal muscle interactome centered on proteins involved in muscular dystrophies: LGMD interactome
- PMID: 23414517
- PMCID: PMC3610214
- DOI: 10.1186/2044-5040-3-3
A human skeletal muscle interactome centered on proteins involved in muscular dystrophies: LGMD interactome
Abstract
Background: The complexity of the skeletal muscle and the identification of numerous human disease-causing mutations in its constitutive proteins make it an interesting tissue for proteomic studies aimed at understanding functional relationships of interacting proteins in both health and diseases.
Method: We undertook a large-scale study using two-hybrid screens and a human skeletal-muscle cDNA library to establish a proteome-scale map of protein-protein interactions centered on proteins involved in limb-girdle muscular dystrophies (LGMD). LGMD is a group of more than 20 different neuromuscular disorders that principally affect the proximal pelvic and shoulder girdle muscles.
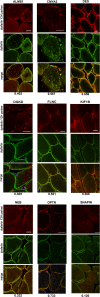
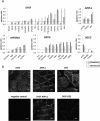
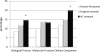
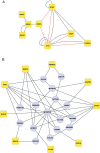
Results and conclusion: The interaction network we unraveled incorporates 1018 proteins connected by 1492 direct binary interactions and includes 1420 novel protein-protein interactions. Computational, experimental and literature-based analyses were performed to assess the overall quality of this network. Interestingly, LGMD proteins were shown to be highly interconnected, in particular indirectly through sarcomeric proteins. In-depth mining of the LGMD-centered interactome identified new candidate genes for orphan LGMDs and other neuromuscular disorders. The data also suggest the existence of functional links between LGMD2B/dysferlin and gene regulation, between LGMD2C/γ-sarcoglycan and energy control and between LGMD2G/telethonin and maintenance of genome integrity. This dataset represents a valuable resource for future functional investigations.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

