Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions
- PMID: 23415234
- PMCID: PMC3616380
- DOI: 10.1016/j.cell.2013.01.042
Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions
Abstract
The bacterial type VI secretion system (T6SS) is a dynamic organelle that bacteria use to target prey cells for inhibition via translocation of effector proteins. Time-lapse fluorescence microscopy has documented striking dynamics of opposed T6SS organelles in adjacent sister cells of Pseudomonas aeruginosa. Such cell-cell interactions have been termed "T6SS dueling" and likely reflect a biological process that is driven by T6SS antibacterial attack. Here, we show that T6SS dueling behavior strongly influences the ability of P. aeruginosa to prey upon heterologous bacterial species. We show that, in the case of P. aeruginosa, T6SS-dependent killing of either Vibrio cholerae or Acinetobacter baylyi is greatly stimulated by T6SS activity occurring in those prey species. Our data suggest that, in P. aeruginosa, T6SS organelle assembly and lethal counterattack are regulated by a signal that corresponds to the point of attack of the T6SS apparatus elaborated by a second aggressive T6SS(+) bacterial cell. PAPERFLICK:
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

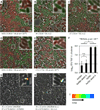


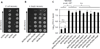
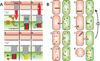
Comment in
-
Bacterial physiology: Ready, aim, fire!Nat Rev Microbiol. 2013 Apr;11(4):224-5. doi: 10.1038/nrmicro3004. Nat Rev Microbiol. 2013. PMID: 23493144 No abstract available.
References
-
- Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, Remers S, Webb J, Braaten BA, Silhavy TJ, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70:323–340. - PMC - PubMed
-
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. - PubMed
-
- Axelrod R, Hamilton WD. The evolution of cooperation. Science. 1981;211:1390–1396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

