Cell-free expressed bacteriorhodopsin in different soluble membrane mimetics: biophysical properties and NMR accessibility
- PMID: 23415558
- PMCID: PMC3595385
- DOI: 10.1016/j.str.2013.01.005
Cell-free expressed bacteriorhodopsin in different soluble membrane mimetics: biophysical properties and NMR accessibility
Abstract
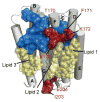
Selecting a suitable membrane-mimicking environment is of fundamental importance for the investigation of membrane proteins. Nonconventional surfactants, such as amphipathic polymers (amphipols) and lipid bilayer nanodiscs, have been introduced as promising environments that may overcome intrinsic disadvantages of detergent micelle systems. However, structural insights into the effects of different environments on the embedded protein are limited. Here, we present a comparative study of the heptahelical membrane protein bacteriorhodopsin in detergent micelles, amphipols, and nanodiscs. Our results confirm that nonconventional environments can increase stability of functional bacteriorhodopsin, and demonstrate that well-folded heptahelical membrane proteins are, in principle, accessible by solution-NMR methods in amphipols and phospholipid nanodiscs. Our data distinguish regions of bacteriorhodopsin that mediate membrane/solvent contacts in the tested environments, whereas the protein's functional inner core remains almost unperturbed. The presented data allow comparing the investigated membrane mimetics in terms of NMR spectral quality and thermal stability required for structural studies.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures



References
-
- Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–222. - PubMed
-
- Bermel W, Bertini I, Felli IC, Peruzzini R, Pierattelli R. Exclusively heteronuclear NMR experiments to obtain structural and dynamic information on proteins. Chemphyschem. 2010;11:689–695. - PubMed
-
- Brouillette CG, McMichens RB, Stern LJ, Khorana HG. Structure and thermal stability of monomeric bacteriorhodopsin in mixed phospholipid/detergent micelles. Proteins. 1989;5:38–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

