DEAD-box helicases as integrators of RNA, nucleotide and protein binding
- PMID: 23416748
- PMCID: PMC3661757
- DOI: 10.1016/j.bbagrm.2013.02.002
DEAD-box helicases as integrators of RNA, nucleotide and protein binding
Abstract
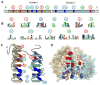
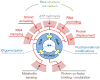
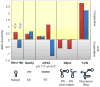
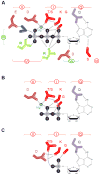
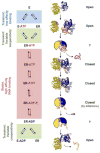
DEAD-box helicases perform diverse cellular functions in virtually all steps of RNA metabolism from Bacteria to Humans. Although DEAD-box helicases share a highly conserved core domain, the enzymes catalyze a wide range of biochemical reactions. In addition to the well established RNA unwinding and corresponding ATPase activities, DEAD-box helicases promote duplex formation and displace proteins from RNA. They can also function as assembly platforms for larger ribonucleoprotein complexes, and as metabolite sensors. This review aims to provide a perspective on the diverse biochemical features of DEAD-box helicases and connections to structural information. We discuss these data in the context of a model that views the enzymes as integrators of RNA, nucleotide, and protein binding. This article is part of a Special Issue entitled: The Biology of RNA helicases - Modulation for life.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures
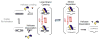
References
-
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. - PubMed
-
- Linder P. The Dynamic Life with DEAD-box RNA Helicases. In: Jankowsky E, editor. RNA Helicases: RSC Biomolecular Sciences. Vol. 19. 2010. pp. 32–60.
-
- Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

