Transcription-dependent dynamic supercoiling is a short-range genomic force
- PMID: 23416947
- PMCID: PMC3594045
- DOI: 10.1038/nsmb.2517
Transcription-dependent dynamic supercoiling is a short-range genomic force
Abstract
Transcription has the capacity to mechanically modify DNA topology, DNA structure and nucleosome arrangement. Resulting from ongoing transcription, these modifications in turn may provide instant feedback to the transcription machinery. To substantiate the connection between transcription and DNA dynamics, we charted an ENCODE map of transcription-dependent dynamic supercoiling in human Burkitt's lymphoma cells by using psoralen photobinding to probe DNA topology in vivo. Dynamic supercoils spread ~1.5 kilobases upstream of the start sites of active genes. Low- and high-output promoters handled this torsional stress differently, as shown by using inhibitors of transcription and topoisomerases and by chromatin immunoprecipation of RNA polymerase and topoisomerases I and II. Whereas lower outputs are managed adequately by topoisomerase I, high-output promoters additionally require topoisomerase II. The genome-wide coupling between transcription and DNA topology emphasizes the importance of dynamic supercoiling for gene regulation.
Figures


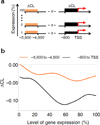
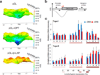

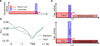
Comment in
-
Genome organization: Supercoiled roles.Nat Rev Genet. 2013 Apr;14(4):238-9. doi: 10.1038/nrg3452. Epub 2013 Mar 5. Nat Rev Genet. 2013. PMID: 23458857 No abstract available.
-
The helix turns at 60: writhing free in chromosomes.Nat Struct Mol Biol. 2013 Mar;20(3):251-3. doi: 10.1038/nsmb.2533. Nat Struct Mol Biol. 2013. PMID: 23463308 No abstract available.
References
-
- Lavelle C. Forces and torques in the nucleus: chromatin under mechanical constraints. Biochem Cell Biol. 2009;87(1):307–322. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

