CLIC4 regulates TGF-β-dependent myofibroblast differentiation to produce a cancer stroma
- PMID: 23416981
- PMCID: PMC3912213
- DOI: 10.1038/onc.2013.18
CLIC4 regulates TGF-β-dependent myofibroblast differentiation to produce a cancer stroma
Abstract
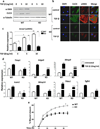
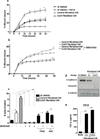
Cancer stroma has a profound influence on tumor development and progression. The conversion of fibroblasts to activated myofibroblasts is a hallmark of reactive tumor stroma. Among a number of factors involved in this conversion, transforming growth factor (TGF)-β has emerged as a major regulator. CLIC4, an integral protein in TGF-β signaling, is highly upregulated in stroma of multiple human cancers, and overexpression of CLIC4 in stromal cells enhances the growth of cancer xenografts. In this study, we show that conditioned media from tumor cell lines induces expression of both CLIC4 and the myofibroblast marker alpha smooth muscle actin (α-SMA) in stromal fibroblasts via TGF-β signaling. Genetic ablation of CLIC4 in primary fibroblasts prevents or reduces constitutive or TGF-β-induced expression of α-SMA and extracellular matrix components that are markers of myofibroblasts. CLIC4 is required for the activation of p38 map kinase by TGF-β, a pathway that signals myofibroblast conversion in stromal cells. This requirement involves the interaction of CLIC4 with PPM1a, the selective phosphatase of activated p38. Conditioned media from fibroblasts overexpressing CLIC4 increases tumor cell migration and invasion in a TGF-β-dependent manner and promotes epithelial to mesenchymal transition indicating that high stromal CLIC4 serves to enhance tumor invasiveness and progression. Thus, CLIC4 is significantly involved in the development of a nurturing tumor microenvironment by enhancing TGF-β signaling in a positive feedback loop. Targeting CLIC4 in tumor stroma should be considered as a strategy to mitigate some of the tumor enhancing effects of the cancer stroma.
Conflict of interest statement
The authors have no conflicts of interest.
Figures



References
-
- Suh KS, Yuspa SH. Intracellular chloride channels: critical mediators of cell viability and potential targets for cancer therapy. Curr Pharm Des. 2005;11(21):2753–2764. - PubMed
-
- Littler DR, Harrop SJ, Goodchild SC, Phang JM, Mynott AV, Jiang L, et al. The enigma of the CLIC proteins: Ion channels, redox proteins, enzymes, scaffolding proteins? FEBS Lett. 2010 May 17;584(10):2093–2101. - PubMed
-
- Littler DR, Harrop SJ, Fairlie WD, Brown LJ, Pankhurst GJ, Pankhurst S, et al. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J Biol Chem. 2004 Mar 5;279(10):9298–9305. - PubMed
-
- Littler DR, Assaad NN, Harrop SJ, Brown LJ, Pankhurst GJ, Luciani P, et al. Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4. FEBS J. 2005 Oct;272(19):4996–5007. - PubMed
-
- Shorning BY, Wilson DB, Meehan RR, Ashley RH. Molecular cloning and developmental expression of two Chloride Intracellular Channel (CLIC) genes in Xenopus laevis. Dev Genes Evol. 2003 Oct;213(10):514–518. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

