PRoNTo: pattern recognition for neuroimaging toolbox
- PMID: 23417655
- PMCID: PMC3722452
- DOI: 10.1007/s12021-013-9178-1
PRoNTo: pattern recognition for neuroimaging toolbox
Abstract
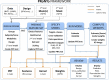
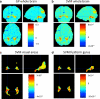
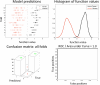
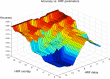
In the past years, mass univariate statistical analyses of neuroimaging data have been complemented by the use of multivariate pattern analyses, especially based on machine learning models. While these allow an increased sensitivity for the detection of spatially distributed effects compared to univariate techniques, they lack an established and accessible software framework. The goal of this work was to build a toolbox comprising all the necessary functionalities for multivariate analyses of neuroimaging data, based on machine learning models. The "Pattern Recognition for Neuroimaging Toolbox" (PRoNTo) is open-source, cross-platform, MATLAB-based and SPM compatible, therefore being suitable for both cognitive and clinical neuroscience research. In addition, it is designed to facilitate novel contributions from developers, aiming to improve the interaction between the neuroimaging and machine learning communities. Here, we introduce PRoNTo by presenting examples of possible research questions that can be addressed with the machine learning framework implemented in PRoNTo, and cannot be easily investigated with mass univariate statistical analysis.
Figures



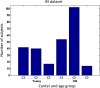
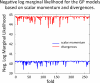
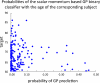
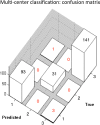
References
-
- Bishop, C. (2006). Pattern recognition and machine learning. Springer.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

