The KMN protein network--chief conductors of the kinetochore orchestra
- PMID: 23418356
- PMCID: PMC3585512
- DOI: 10.1242/jcs.093724
The KMN protein network--chief conductors of the kinetochore orchestra
Abstract
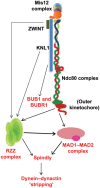
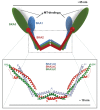
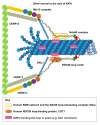
Successful completion of mitosis requires that sister kinetochores become attached end-on to the plus ends of spindle microtubules (MTs) in prometaphase, thereby forming kinetochore microtubules (kMTs) that tether one sister to one spindle pole and the other sister to the opposite pole. Sites for kMT attachment provide at least four key functions: robust and dynamic kMT anchorage; force generation that can be coupled to kMT plus-end dynamics; correction of errors in kMT attachment; and control of the spindle assembly checkpoint (SAC). The SAC typically delays anaphase until chromosomes achieve metaphase alignment with each sister kinetochore acquiring a full complement of kMTs. Although it has been known for over 30 years that MT motor proteins reside at kinetochores, a highly conserved network of protein complexes, called the KMN network, has emerged in recent years as the primary interface between the kinetochore and kMTs. This Commentary will summarize recent advances in our understanding of the role of the KMN network for the key kinetochore functions, with a focus on human cells.
Figures
References
-
- Bolanos–Garcia V. M., Lischetti T., Matak–Vinković D., Cota E., Simpson P. J., Chirgadze D. Y., Spring D. R., Robinson C. V., Nilsson J., Blundell T. L. (2011). Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding Site. Structure 19, 1691–1700 10.1016/j.str.2011.09.017 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

