Deregulated Nras expression in knock-in animals harboring a gammaretroviral long terminal repeat at the Nras/Csde1 locus
- PMID: 23418499
- PMCID: PMC3572152
- DOI: 10.1371/journal.pone.0056029
Deregulated Nras expression in knock-in animals harboring a gammaretroviral long terminal repeat at the Nras/Csde1 locus
Abstract
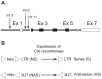
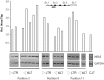
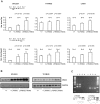
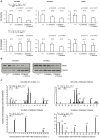
To investigate mechanisms and phenotypic effects of insertional mutagenesis by gammaretroviruses, we have developed mouse lines containing a single Akv 1-99 long terminal repeat (LTR) and a floxed PGK/Tn5 neomycin cassette at the Nras proto-oncogene at positions previously identified as viral integration sites in Akv 1-99 induced tumors. The insert did not compromise the embryonic development, however, the cassette had an effect on Nras expression in all tissues analyzed. Cre-mediated excision of the PGK/Tn5 neomycin cassette in two of the lines caused upregulation of Nras. Altogether, the knock-in alleles are characterized by modulation of expression of the target gene from more than ten-fold upregulation to three-fold downregulation and exemplify various mechanisms of deregulation by insertional mutagenesis. LTR knock-in mice may serve as a tool to investigate mechanisms of retroviral insertional mutagenesis and as a way of constitutive or induced modulation of expression of a target gene.
Conflict of interest statement
Figures

References
-
- Uren AG, Kool J, Berns A, van Lohuizen M (2005) Retroviral insertional mutagenesis: past, present and future. Oncogene 24: 7656–7672. - PubMed
-
- Blyth K, Vaillant F, Hanlon L, Mackay N, Bell M, et al. (2006) Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res 66: 2195–2201. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

