A highly-conserved single-stranded DNA-binding protein in Xanthomonas functions as a harpin-like protein to trigger plant immunity
- PMID: 23418541
- PMCID: PMC3571957
- DOI: 10.1371/journal.pone.0056240
A highly-conserved single-stranded DNA-binding protein in Xanthomonas functions as a harpin-like protein to trigger plant immunity
Abstract
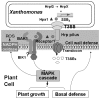
Harpins are produced by gram-negative phytopathogenic bacteria and typically elicit hypersensitive response (HR) in non-host plants. The characterization of harpins in Xanthomonas species is largely unexplored. Here we demonstrate that Xanthomonas produce a highly conserved single-stranded DNA-binding protein (SSB(X)) that elicits HR in tobacco as by harpin Hpa1. SSB(X), like Hpa1, is an acidic, glycine-rich, heat-stable protein that lacks cysteine residues. SSB(X)-triggered HR in tobacco, as by Hpa1, is characterized by the oxidative burst, the expression of HR markers (HIN1, HSR203J), pathogenesis-related genes, and callose deposition. Both SSB(X)- and Hpa1-induced HRs can be inhibited by general metabolism inhibitors actinomycin D, cycloheximide, and lanthanum chloride. Furthermore, those HRs activate the expression of BAK1 and BIK1 genes that are essential for induction of mitogen-activated protein kinase (MAPK) and salicylic acid pathways. Once applied to plants, SSB(X) induces resistance to the fungal pathogen Alternaria alternata and enhances plant growth. When ssb(X)was deleted in X. oryzae pv. oryzicola, the causal agent of bacterial leaf streak in rice, the resulting ssb(Xoc)mutant was reduced in virulence and bacterial growth in planta, but retained its ability to trigger HR in tobacco. Interestingly, ssb(Xoc)contains an imperfect PIP-box (plant-inducible promoter) and the expression of ssb(Xoc)is regulated by HrpX, which belongs to the AraC family of transcriptional activators. Immunoblotting evidence showed that SSB(x) secretion requires a functional type-III secretion system as Hpa1 does. This is the first report demonstrating that Xanthomonas produce a highly-conserved SSB(X) that functions as a harpin-like protein for plant immunity.
Conflict of interest statement
Figures







References
-
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329. - PubMed
-
- Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465. - PubMed
-
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276. - PubMed
-
- Felix G, Regenass M, Boller T (1993) Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J 4: 307–316.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

