Multifactor dimensionality reduction reveals a three-locus epistatic interaction associated with susceptibility to pulmonary tuberculosis
- PMID: 23418869
- PMCID: PMC3618340
- DOI: 10.1186/1756-0381-6-4
Multifactor dimensionality reduction reveals a three-locus epistatic interaction associated with susceptibility to pulmonary tuberculosis
Abstract
Background: Identifying high-order genetics associations with non-additive (i.e. epistatic) effects in population-based studies of common human diseases is a computational challenge. Multifactor dimensionality reduction (MDR) is a machine learning method that was designed specifically for this problem. The goal of the present study was to apply MDR to mining high-order epistatic interactions in a population-based genetic study of tuberculosis (TB).
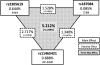
Results: The study used a previously published data set consisting of 19 candidate single-nucleotide polymorphisms (SNPs) in 321 pulmonary TB cases and 347 healthy controls from Guniea-Bissau in Africa. The ReliefF algorithm was applied first to generate a smaller set of the five most informative SNPs. MDR with 10-fold cross-validation was then applied to look at all possible combinations of two, three, four and five SNPs. The MDR model with the best testing accuracy (TA) consisted of SNPs rs2305619, rs187084, and rs11465421 (TA = 0.588) in PTX3, TLR9 and DC-Sign, respectively. A general 1000-fold permutation test of the null hypothesis of no association confirmed the statistical significance of the model (p = 0.008). An additional 1000-fold permutation test designed specifically to test the linear null hypothesis that the association effects are only additive confirmed the presence of non-additive (i.e. nonlinear) or epistatic effects (p = 0.013). An independent information-gain measure corroborated these results with a third-order epistatic interaction that was stronger than any lower-order associations.
Conclusions: We have identified statistically significant evidence for a three-way epistatic interaction that is associated with susceptibility to TB. This interaction is stronger than any previously described one-way or two-way associations. This study highlights the importance of using machine learning methods that are designed to embrace, rather than ignore, the complexity of common diseases such as TB. We recommend future studies of the genetics of TB take into account the possibility that high-order epistatic interactions might play an important role in disease susceptibility.
Figures

References
-
- De Wit E, van der Merwe L, van Helden PD, Hoal EG. Gene-gene interaction between tuberculosis candidate genes in a South African population. Mamm Genome. 2011;22(1–2):100–110. - PubMed
-
- Olesen R, Wejse C, Velez DR, Bisseye C, Sodemann M, Aaby P, Rabna P, Worwui A, Chapman H, Diatta M, Adegbola RA, Hill PC, Østergaard L, Williams SM, Sirugo G. DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes Immun. 2007;8(suppl 6):456–467. - PubMed
-
- Moore JH, Gilbert JC, Tsai C, Chiang F, Holden T, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241:252–261. doi: 10.1016/j.jtbi.2005.11.036. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

