Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance
- PMID: 23419278
- PMCID: PMC3705945
- DOI: 10.1038/nrg3351
Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance
Abstract
The evolution of antibiotic resistance can now be rapidly tracked with high-throughput technologies for bacterial genotyping and phenotyping. Combined with new approaches to evolve resistance in the laboratory and to characterize clinically evolved resistant pathogens, these methods are revealing the molecular basis and rate of evolution of antibiotic resistance under treatment regimens of single drugs or drug combinations. In this Progress article, we review these new tools for studying the evolution of antibiotic resistance and discuss how the genomic and evolutionary insights they provide could transform the diagnosis, treatment and predictability of antibiotic resistance in bacterial infections.
Figures
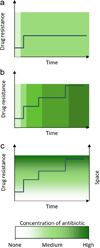
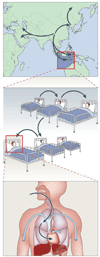

References
-
- The evolving threat of antimicrobial resistance: Options for action. Geneva: World Health Organization; 2012.
-
- Paterson DL. Resistance in gram-negative bacteria: enterobacteriaceae. Am J Med. 2006;119:S20–S28. discussion S62-70. - PubMed
-
- Morar M, Wright GD. The genomic enzymology of antibiotic resistance. Annu Rev Genet. 2010;44:25–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

