Minimal residual disease quantification using consensus primers and high-throughput IGH sequencing predicts post-transplant relapse in chronic lymphocytic leukemia
- PMID: 23419792
- PMCID: PMC3740398
- DOI: 10.1038/leu.2013.52
Minimal residual disease quantification using consensus primers and high-throughput IGH sequencing predicts post-transplant relapse in chronic lymphocytic leukemia
Abstract
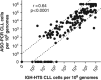
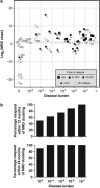
Quantification of minimal residual disease (MRD) following allogeneic hematopoietic cell transplantation (allo-HCT) predicts post-transplant relapse in patients with chronic lymphocytic leukemia (CLL). We utilized an MRD-quantification method that amplifies immunoglobulin heavy chain (IGH) loci using consensus V and J segment primers followed by high-throughput sequencing (HTS), enabling quantification with a detection limit of one CLL cell per million mononuclear cells. Using this IGH-HTS approach, we analyzed MRD patterns in over 400 samples from 40 CLL patients who underwent reduced-intensity allo-HCT. Nine patients relapsed within 12 months post-HCT. Of the 31 patients in remission at 12 months post-HCT, disease-free survival was 86% in patients with MRD <10(-4) and 20% in those with MRD ≥10(-4) (relapse hazard ratio (HR) 9.0; 95% confidence interval (CI) 2.5-32; P<0.0001), with median follow-up of 36 months. Additionally, MRD predicted relapse at other time points, including 9, 18 and 24 months post-HCT. MRD doubling time <12 months with disease burden ≥10(-5) was associated with relapse within 12 months of MRD assessment in 50% of patients, and within 24 months in 90% of patients. This IGH-HTS method may facilitate routine MRD quantification in clinical trials.
Figures



References
-
- Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. - PMC - PubMed
-
- Rawstron AC, Villamor N, Ritgen M, Bottcher S, Ghia P, Zehnder JL, et al. International standardized approach for flow cytometric residual disease monitoring in chronic lymphocytic leukaemia. Leukemia. 2007;21:956–964. - PubMed
-
- Rawstron AC, Bottcher S, Letestu R, Villamor N, Fazi C, Kartsios H, et al. Improving efficiency and sensitivity: European Research Initiative in CLL (ERIC) update on the international harmonised approach for flow cytometric residual disease monitoring in CLL. Leukemia. 2013;27:142–149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

