Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts
- PMID: 23420232
- PMCID: PMC3638215
- DOI: 10.7326/0003-4819-158-4-201302190-00003
Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts
Abstract
Chinese translation
Background: Hepatitis C virus (HCV) infections occur worldwide and either spontaneously resolve or persist and markedly increase the person's lifetime risk for cirrhosis and hepatocellular carcinoma. Although HCV persistence occurs more often in persons of African ancestry and persons with genetic variants near interleukin-28B (IL-28B), the genetic basis is not well-understood.
Objective: To evaluate the host genetic basis for spontaneous resolution of HCV infection.
Design: 2-stage, genome-wide association study.
Setting: 13 international multicenter study sites.
Patients: 919 persons with serum HCV antibodies but no HCV RNA (spontaneous resolution) and 1482 persons with serum HCV antibodies and HCV RNA (persistence).
Measurements: Frequencies of 792 721 single nucleotide polymorphisms (SNPs).
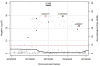
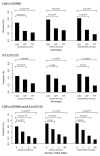
Results: Differences in allele frequencies between persons with spontaneous resolution and persistence were identified on chromosomes 19q13.13 and 6p21.32. On chromosome 19, allele frequency differences localized near IL-28B and included rs12979860 (overall per-allele OR, 0.45; P = 2.17 × 10-30) and 10 additional SNPs spanning 55 000 base pairs. On chromosome 6, allele frequency differences localized near genes for HLA class II and included rs4273729 (overall per-allele OR, 0.59; P = 1.71 × 10-16) near DQB1*03:01 and an additional 116 SNPs spanning 1 090 000 base pairs. The associations in chromosomes 19 and 6 were independent and additive and explain an estimated 14.9% (95% CI, 8.5% to 22.6%) and 15.8% (CI, 4.4% to 31.0%) of the variation in HCV resolution in persons of European and African ancestry, respectively. Replication of the chromosome 6 SNP, rs4272729, in an additional 745 persons confirmed the findings (P = 0.015).
Limitation: Epigenetic effects were not studied.
Conclusion: IL-28B and HLA class II are independently associated with spontaneous resolution of HCV infection, and SNPs marking IL-28B and DQB1*03:01 may explain approximately 15% of spontaneous resolution of HCV infection.
Figures



References
-
- Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–7. - PubMed
-
- Villano SA, Vlahov D, Nelson KE, Cohn S, Thomas DL. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology. 1999;29:908–14. - PubMed
-
- Kenny-Walsh E. Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. Irish Hepatology Research Group. New England Journal of Medicine. 1999;340:1228–33. - PubMed
-
- Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UO1-AI-34994/AI/NIAID NIH HHS/United States
- UO1-AI-34989/AI/NIAID NIH HHS/United States
- AI082630/AI/NIAID NIH HHS/United States
- R01HL076902/HL/NHLBI NIH HHS/United States
- U19AI088791/AI/NIAID NIH HHS/United States
- UO1-AI-35041/AI/NIAID NIH HHS/United States
- UO1-AI-35004/AI/NIAID NIH HHS/United States
- U19 AI082630/AI/NIAID NIH HHS/United States
- UO1-AI-34993/AI/NIAID NIH HHS/United States
- U19 AI066345/AI/NIAID NIH HHS/United States
- UL1-RR025005/RR/NCRR NIH HHS/United States
- DA12568/DA/NIDA NIH HHS/United States
- UL1-RR024996/RR/NCRR NIH HHS/United States
- UO1-AI-42590/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- UO1-AI-31834/AI/NIAID NIH HHS/United States
- U01 DA036297/DA/NIDA NIH HHS/United States
- UO1-HD-32632/HD/NICHD NIH HHS/United States
- R01 DA033541/DA/NIDA NIH HHS/United States
- UO1-AI-35042/AI/NIAID NIH HHS/United States
- R01-DA16159/DA/NIDA NIH HHS/United States
- P30 DK026743/DK/NIDDK NIH HHS/United States
- R01 DA013324/DA/NIDA NIH HHS/United States
- UO1-AI-35040/AI/NIAID NIH HHS/United States
- DA04334/DA/NIDA NIH HHS/United States
- R01-DA21550/DA/NIDA NIH HHS/United States
- R01013324/PHS HHS/United States
- UO1-AI-35039/AI/NIAID NIH HHS/United States
- R01 DA012568/DA/NIDA NIH HHS/United States
- UL1 TR000457/TR/NCATS NIH HHS/United States
- G1001738/MRC_/Medical Research Council/United Kingdom
- HHSN261200800001E/CA/NCI NIH HHS/United States
- DA033541/DA/NIDA NIH HHS/United States
- UO1-AI-35043/AI/NIAID NIH HHS/United States
- U19 AI088791/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
