A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin-tropomyosin
- PMID: 23420843
- PMCID: PMC3617264
- DOI: 10.1074/jbc.M113.451161
A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin-tropomyosin
Abstract
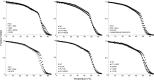
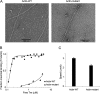
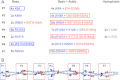
Actin filament cytoskeletal and muscle functions are regulated by actin binding proteins using a variety of mechanisms. A universal actin filament regulator is the protein tropomyosin, which binds end-to-end along the length of the filament. The actin-tropomyosin filament structure is unknown, but there are atomic models in different regulatory states based on electron microscopy reconstructions, computational modeling of actin-tropomyosin, and docking of atomic resolution structures of tropomyosin to actin filament models. Here, we have tested models of the actin-tropomyosin interface in the "closed state" where tropomyosin binds to actin in the absence of myosin or troponin. Using mutagenesis coupled with functional analyses, we determined residues of actin and tropomyosin required for complex formation. The sites of mutations in tropomyosin were based on an evolutionary analysis and revealed a pattern of basic and acidic residues in the first halves of the periodic repeats (periods) in tropomyosin. In periods P1, P4, and P6, basic residues are most important for actin affinity, in contrast to periods P2, P3, P5, and P7, where both basic and acidic residues or predominantly acidic residues contribute to actin affinity. Hydrophobic interactions were found to be relatively less important for actin binding. We mutated actin residues in subdomains 1 and 3 (Asp(25)-Glu(334)-Lys(326)-Lys(328)) that are poised to make electrostatic interactions with the residues in the repeating motif on tropomyosin in the models. Tropomyosin failed to bind mutant actin filaments. Our mutagenesis studies provide the first experimental support for the atomic models of the actin-tropomyosin interface.
Figures



References
-
- Gunning P., O'Neill G., Hardeman E. (2008) Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 88, 1–35 - PubMed
-
- Kee A. J., Hardeman E. C. (2008) Tropomyosins in skeletal muscle diseases. Adv. Exp. Med. Biol. 644, 143–157 - PubMed
-
- Wieczorek D. F., Jagatheesan G., Rajan S. (2008) The role of tropomyosin in heart disease. Adv. Exp. Med. Biol. 644, 132–142 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

