A new way to degrade heme: the Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO
- PMID: 23420845
- PMCID: PMC3617252
- DOI: 10.1074/jbc.M112.448399
A new way to degrade heme: the Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO
Abstract
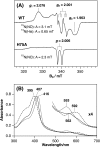
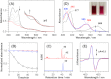
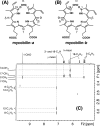
MhuD is an oxygen-dependent heme-degrading enzyme from Mycobacterium tuberculosis with high sequence similarity (∼45%) to Staphylococcus aureus IsdG and IsdI. Spectroscopic and mutagenesis studies indicate that the catalytically active 1:1 heme-MhuD complex has an active site structure similar to those of IsdG and IsdI, including the nonplanarity (ruffling) of the heme group bound to the enzyme. Distinct from the canonical heme degradation, we have found that the MhuD catalysis does not generate CO. Product analyses by electrospray ionization-MS and NMR show that MhuD cleaves heme at the α-meso position but retains the meso-carbon atom at the cleavage site, which is removed by canonical heme oxygenases. The novel tetrapyrrole product of MhuD, termed "mycobilin," has an aldehyde group at the cleavage site and a carbonyl group at either the β-meso or the δ-meso position. Consequently, MhuD catalysis does not involve verdoheme, the key intermediate of ring cleavage by canonical heme oxygenase enzymes. Ruffled heme is apparently responsible for the heme degradation mechanism unique to MhuD. In addition, MhuD heme degradation without CO liberation is biologically significant as one of the signals of M. tuberculosis transition to dormancy is mediated by the production of host CO.
Figures






References
-
- Maines M. D. (1997) The heme oxygenase system: A regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 37, 517–554 - PubMed
-
- Yoshida T., Kikuchi G. (1978) Features of the reaction of heme degradation catalyzed by the reconstituted microsomal heme oxygenase system. J. Biol. Chem. 253, 4230–4236 - PubMed
-
- Tenhunen R., Marver H. S., Schmid R. (1969) Microsomal heme oxygenase: characterization of the enzyme. J. Biol. Chem. 244, 6388–6394 - PubMed
-
- Ortiz de Montellano P. R. (1998) Heme oxygenase mechanism: Evidence for an electrophilic, ferric peroxide species. Acc. Chem. Res. 31, 543–549
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

