Grb2 controls phosphorylation of FGFR2 by inhibiting receptor kinase and Shp2 phosphatase activity
- PMID: 23420874
- PMCID: PMC3575544
- DOI: 10.1083/jcb.201204106
Grb2 controls phosphorylation of FGFR2 by inhibiting receptor kinase and Shp2 phosphatase activity
Abstract
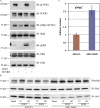
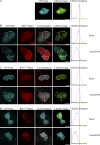
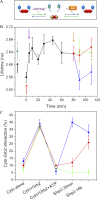
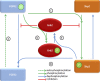
Constitutive receptor tyrosine kinase phosphorylation requires regulation of kinase and phosphatase activity to prevent aberrant signal transduction. A dynamic mechanism is described here in which the adaptor protein, growth factor receptor-bound protein 2 (Grb2), controls fibroblast growth factor receptor 2 (FGFR2) signaling by regulating receptor kinase and SH2 domain-containing protein tyrosine phosphatase 2 (Shp2) phosphatase activity in the absence of extracellular stimulation. FGFR2 cycles between its kinase-active, partially phosphorylated, nonsignaling state and its Shp2-dephosphorylated state. Concurrently, Shp2 cycles between its FGFR2-phosphorylated and dephosphorylated forms. Both reciprocal activities of FGFR2 and Shp2 were inhibited by binding of Grb2 to the receptor. Phosphorylation of Grb2 by FGFR2 abrogated its binding to the receptor, resulting in up-regulation of both FGFR2's kinase and Shp2's phosphatase activity. Dephosphorylation of Grb2 by Shp2 rescued the FGFR2-Grb2 complex. This cycling of enzymatic activity results in a homeostatic, signaling-incompetent state. Growth factor binding perturbs this background cycling, promoting increased FGFR2 phosphorylation and kinase activity, Grb2 dissociation, and downstream signaling. Grb2 therefore exerts constitutive control over the mutually dependent activities of FGFR2 and Shp2.
Figures




References
-
- Agazie Y.M., Hayman M.J. 2003. Development of an efficient “substrate-trapping” mutant of Src homology phosphotyrosine phosphatase 2 and identification of the epidermal growth factor receptor, Gab1, and three other proteins as target substrates. J. Biol. Chem. 278:13952–13958 10.1074/jbc.M210670200 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

