Concentration-dependent activity of antibiotics in natural environments
- PMID: 23422936
- PMCID: PMC3574975
- DOI: 10.3389/fmicb.2013.00020
Concentration-dependent activity of antibiotics in natural environments
Abstract
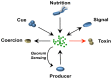
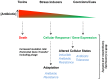
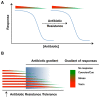
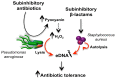
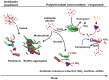
Bacterial responses to antibiotics are concentration-dependent. At high concentrations, antibiotics exhibit antimicrobial activities on susceptible cells, while subinhibitory concentrations induce diverse biological responses in bacteria. At non-lethal concentrations, bacteria may sense antibiotics as extracellular chemicals to trigger different cellular responses, which may include an altered antibiotic resistance/tolerance profile. In natural settings, microbes are typically in polymicrobial communities and antibiotic-mediated interactions between species may play a significant role in bacterial community structure and function. However, these aspects have not yet fully been explored at the community level. Here we discuss the different types of interactions mediated by antibiotics and non-antibiotic metabolites as a function of their concentrations and speculate on how these may amplify the overall antibiotic resistance/tolerance and the spread of antibiotic resistance determinants in a context of polymicrobial community.
Keywords: antibiotic; community; cue; interaction; resistance; signal; stress; tolerance.
Figures
References
-
- Aertsen A., Michiels C. W. (2006). Upstream of the SOS response: figure out the trigger. Trends Microbiol. 14 421–423 - PubMed
-
- Allen H. K., Donato J., Wang H. H., Cloud-Hansen K. A., Davies J., Handelsman J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8 251–259 - PubMed
-
- Aminov R. I. (2009). The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 11 2970–2988 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

