Mitotic spindle scaling during Xenopus development by kif2a and importin α
- PMID: 23425906
- PMCID: PMC3576809
- DOI: 10.7554/eLife.00290
Mitotic spindle scaling during Xenopus development by kif2a and importin α
Abstract
Early development of many animals is characterized by rapid cleavages that dramatically decrease cell size, but how the mitotic spindle adapts to changing cell dimensions is not understood. To identify mechanisms that scale the spindle during Xenopus laevis embryogenesis, we established an in vitro system using cytoplasmic extracts prepared from embryos that recapitulates in vivo spindle size differences between stage 3 (4 cells, 37 µm) and stage 8 (∼4000 cells, 18 µm). We identified the kinesin-13 kif2a as a driver of developmental spindle scaling whose microtubule-destabilizing activity is inhibited in stage 3 spindles by the transport receptor importin α, and activated in stage 8 when importin α partitions to a membrane pool. Altering spindle size in developing embryos impaired spindle orientation during metaphase, but chromosome segregation remained robust. Thus, spindle size in Xenopus development is coupled to cell size through a ratiometric mechanism controlling microtubule destabilization.DOI:http://dx.doi.org/10.7554/eLife.00290.001.
Keywords: Xenopus; embryogenesis; intracellular scaling; kif2a; microtubule; mitotic spindle.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
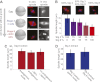

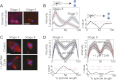

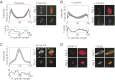


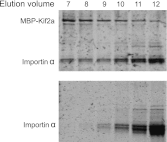

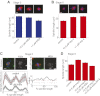
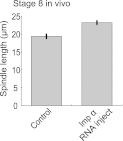
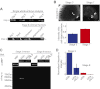
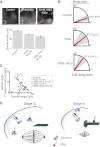
References
-
- Brugues J, Nuzzo V, Mazur E, Needleman DJ. 2012. Nucleation and transport organize microtubules in metaphase spindles. Cell 149:554–64 - PubMed
-
- Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E. 2001. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat Cell Biol 3:228–34 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

