Phosphorylation of lipin 1 and charge on the phosphatidic acid head group control its phosphatidic acid phosphatase activity and membrane association
- PMID: 23426360
- PMCID: PMC3617293
- DOI: 10.1074/jbc.M112.441493
Phosphorylation of lipin 1 and charge on the phosphatidic acid head group control its phosphatidic acid phosphatase activity and membrane association
Abstract
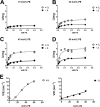
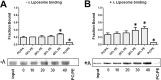
The lipin gene family encodes a class of Mg(2+)-dependent phosphatidic acid phosphatases involved in the de novo synthesis of phospholipids and triglycerides. Unlike other enzymes in the Kennedy pathway, lipins are not integral membrane proteins, and they need to translocate from the cytosol to intracellular membranes to participate in glycerolipid synthesis. The movement of lipin 1 within the cell is closely associated with its phosphorylation status. Although cellular analyses have demonstrated that highly phosphorylated lipin 1 is enriched in the cytosol and dephosphorylated lipin 1 is found on membranes, the effects of phosphorylation on lipin 1 activity and binding to membranes has not been recapitulated in vitro. Herein we describe a new biochemical assay for lipin 1 using mixtures of phosphatidic acid (PA) and phosphatidylethanolamine that reflects its physiological activity and membrane interaction. This depends on our observation that lipin 1 binding to PA in membranes is highly responsive to the electrostatic charge of PA. The studies presented here demonstrate that phosphorylation regulates the ability of the polybasic domain of lipin 1 to recognize di-anionic PA and identify mTOR as a crucial upstream signaling component regulating lipin 1 phosphorylation. These results demonstrate how phosphorylation of lipin 1 together with pH and membrane phospholipid composition play important roles in the membrane association of lipin 1 and thus the regulation of its enzymatic activity.
Figures






References
-
- Péterfy M., Phan J., Xu P., Reue K. (2001) Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27, 121–124 - PubMed
-
- Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., Lawrence J. C., Jr. (2007) Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282, 277–286 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

